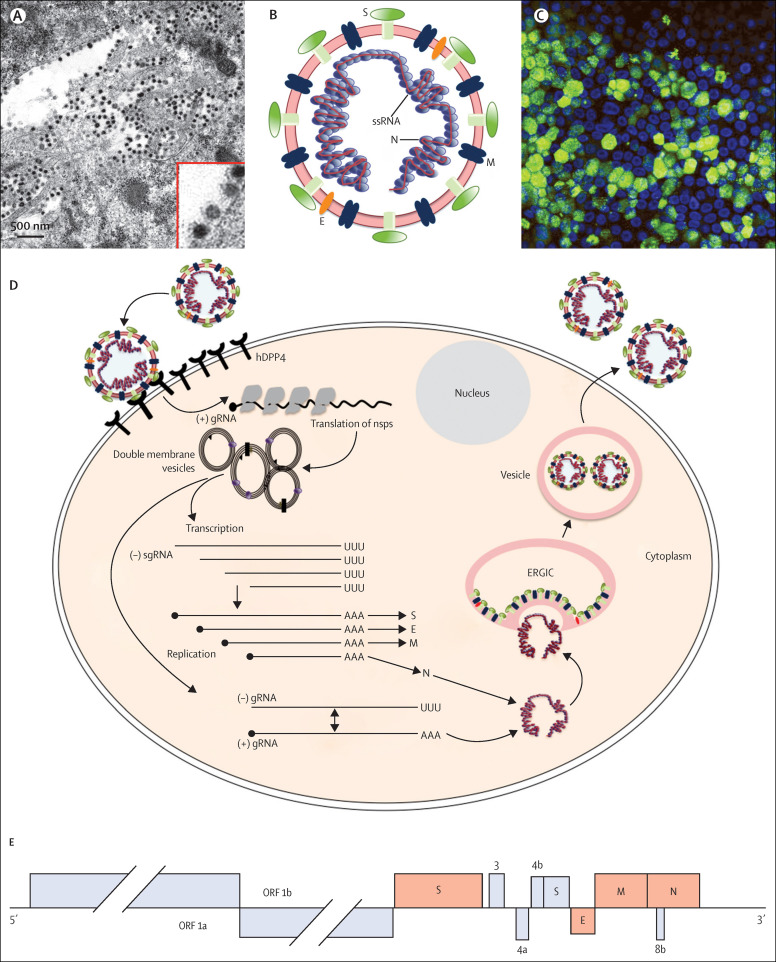
Figure 2.
MERS-CoV virion, replication strategy, and genomic structure
(A) Electron micrograph of MERS-CoV virions in large single membrane vesicles and in the periphery of the cell. Virion spikes are visible as is the electron-dense core consisting of the RNA genome encapsidated in the N protein. Provided by Montserrat Barcena, Ronald Limpens and Eric Snijder (Leiden University Medical Center, Netherlands). (B) MERS-CoV, showing viral RNA structural proteins spike (S), envelope (E), matrix (M), and nucleocapsid (N). (C) Human airway cells infected with MERS-CoV and assessed for viral antigen by indirect immunofluorescence assay with an anti-MERS-CoV N antibody. Green shows MERS-CoV-positive cells, blue shows nuclear stain. A similar assay is used for serological diagnosis of MERS. Provided by Christine Wohlford-Lenane (University of Iowa, Iowa City, IA). (D) Virus life-cycle. (E) The MERS-CoV genome consists of 11 ORFs that code for the virus replication machinery (ORF 1a and ORF 1b) and the major structural proteins: spike (S), envelope (E), matrix (M), and nucleocapsid (N). ORF 1b, produced by a–1 basepair frameshift from ORF 1a, encodes the RNA-dependent RNA polymerase (nsp12), helicase (nsp13), N7-methyltransferase (nsp14), a 3'–5' exonuclease for RNA proofreading (nsp14), 2'-O-methyltransferase (nsp16), and an endonuclease specific for U and found only in nidoviruses (nsp15). MERS-CoV also encodes five accessory proteins in the 3' end of the genome (3a/b, 4, 5, 8b), which share no homology with proteins from host cells or other viruses, including coronaviruses. 4a and 4b are interferon antagonists16, 17 but the functions of the other accessory proteins are unknown.