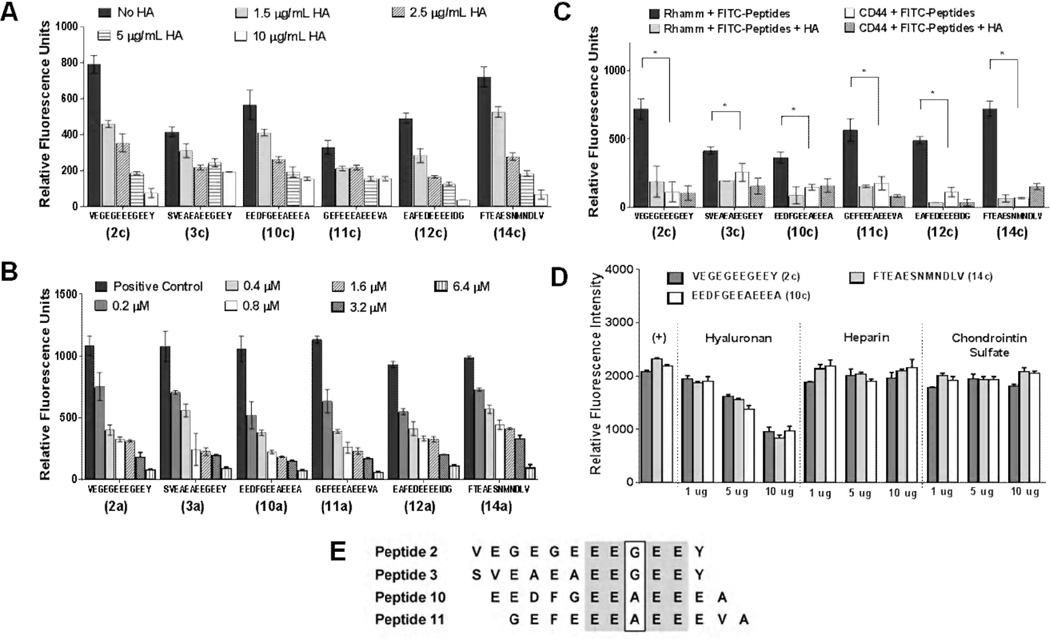
Figure 4. Binding properties of ligand candidates.
(A) Competitive displacement of six selected fluorescein-labelled peptides (2c, 3c, 10c, 11c, 12c and 14c) by HA (220 kDA) in different concentrations to immobilized recombinant RHAMM; (B) Competitive displacement of dye-labeled HA by non-labeled tubulin-derived peptides (2a, 3a, 10a, 11a, 12a and 14a) at different concentrations using ELISA. (C) ELISA binding assay of FITC-conjugated peptides (2c, 3c, 10c, 11c, 12c and 14c) to recombinant CD44 and RHAMM. The negative control (no immobilized RHAMM or CD44) was subtracted for each measurement. 10 µg/mL of the peptides and HA were used for these assays. (D) The ability of HA to compete with FITC-peptides for binding to recombinant RHAMM was compared to heparin and chondroitin sulfate. Only HA effectively competed with the peptides for binding to RHAMM. Each glycosaminoglycan was competed against FITC-peptides at three different concentrations (1 µg/mL, 5 µg/mL and 10 µg/mL). All data show the mean of three measurements ± S.E.M. in three independent experiments. Significant differences (p < 0.05) are marked by asterisks. (E) Sequence comparison of peptides 2, 3, 10 and 11 using Cobalt Multiple Alignment Tool. Identical sequences are shown in grey. A pentapeptide motif, EEXEE (where X is A or G) is present in these peptides.