Abstract
The ability to switch from yeast to hyphal growth is essential for virulence in Candida albicans. The cell surface is the initial point of contact between the fungus and the host. In this work, a free-gel proteomic strategy based on tryptic digestion of live yeast and hyphae cells and protein identification using LC-MS/MS methodology was used to identify cell surface proteins. Using this strategy, a total of 943 proteins were identified, of which 438 were in yeast and 928 were in hyphae. Of these proteins, 79 were closely related to the organization and biogenesis of the cell wall, including 28 GPI-anchored proteins, such as Hyr1 and Sod5 which were detected exclusively in hyphae, and Als2 and Sap10which were detected only in yeast. A group of 17 proteins of unknown function were subsequently studied by analysis of the corresponding deletion mutants. We found that four new proteins, Pst3, Tos1, Orf19.3060 and Orf19.5352 are involved in cell wall integrity and in C. albicans’ engulfment by macrophages. Moreover, the putative NADH-ubiquinone-related proteins, Ali1, Mci4, Orf19.287 and Orf19.7590, are also involved in osmotic and oxidative resistance, yeast to hypha transition and the ability to damage and invade oral epithelial cells.
Keywords: Candida albicans, Trypsin digestion, Surface proteins, Cell wall integrity, Osmotic stress, Oxidative stress
1. Introduction
The cell wall is the most external structure in fungi, confers organization and form to the cell and protects the organism from physical and osmotic damage. Surface proteins play an important role in the pathogenic process as they are the initial point of contact between the cell and the environment. Candida albicans is an important opportunistic fungus that causes a wide variety of diseases in patients, ranging from the superficial mucocutaneous candidiasis (affecting the nails, skin, and oral and genital mucosae) to life-threatening disseminated infections [1,2]. In patient in critical care, candidemia is the most important fungi disease with a 30% mortality rate [3]. The C. albicans’ cell wall maintains structural integrity and acts as intermediate between the cell and the environment. As the initial point of contact with host cells, the cell wall is an obvious target for development of antifungals and vaccines. It is composed of β-1,3-glucan, β-1,6-glucan, a small percentage of chitin and different wall proteins, most of them covalently attached to β-1,6-glucan linkage through a remnant of glycosylphosphatidylinositol (GPI) anchors [4–6]. The non-glucan-linked proteins traffic to the cell surface by either the classical or alternative secretory pathway [7,8]. These cell wall proteins maintain structural integrity, mediate adherence and/or invasion of host cells, or function as enzymes [5,8]. One of these proteins is Ecm33p, a GPI-linked cell wall protein whose absence affects both yeast cells and hyphal morphology and results in an aberrant wall structure and reduced virulence in vitro and in vivo [9,10]. Another GPI-linked cell wall protein is the secreted yeast wall protein Ywp1p, which is covalently linked to glucans of the wall matrix and has the highest expression during yeast exponential growth. The ywp1Δ/Δ mutant has increased adhesiveness and biofilm formation but no obvious change in growth, morphology or virulence, suggesting that Ywp1p promotes dispersal of yeast form cells in C. albicans [11]. Pir proteins (proteins with internal repeats) are an additional group of C. albicans’ cell wall proteins and are linked directly to β-1,3-glucan [12,13]. PIR1 is an essential gene and its abundance changes in response to environmental conditions [13–15]. Among no covalent attachment proteins, Bgl2p is involved in cell wall biogenesis [16]. It is themajor β-1,3-glycosyltransferase and bgl2Δ/Δ mutants have attenuated virulence inmice. Furthermore, Bgl2 is recognized by IgG antibodies from patients with invasive candidiasis, which has diagnostic and prognostic usefulness [17]. Some secreted proteins, such as secreted aspartyl proteinase (SAP) and phospholipase B (PLB) families, must pass through the cell wall and have been detected there [18]. These proteins are hydrolytic enzymes which enable the organism to break down proteins for nutrition, but their relative contribution to C. albicans’ pathogenicity is controversial [19–23]. In addition, many proteins identified on the surface of C. albicans lack classical secretion signal peptides and are dual function proteins, that function as enzymes or chaperones in the cytoplasm and as adhesins, invasins, or immunogens when expressed on the cell surface. These proteins include glyceraldehyde-3-phosphate dehydrogenase (Tdh3), enolase (Eno1) and heat shock proteins such as Hsp70 [24–26].
C. albicans is able to grow in different morphological forms. The ability to switch between yeast and hypha is necessary for virulence [23,27,28]. Both morphological forms are important during infection. The yeast form probably disseminates via the bloodstream, spreading the organism to different host niches, while the hyphal form is invasive and enables the organism to evade phagocytic cells [29,30]. Consequently, C. albicans expresses distinct cell surface proteins in these stages.
The study of cell surface protein composition of yeast and hypha morphologies and their differences will help to find novel therapeutic targets. In recent years, the response of the cell wall proteome to changes in ambient pH and with respect to yeast to hyphal transition has been investigated [31–35]. These classical proteomic approaches involve several steps based on subcellular fractionation which are time-consuming and laborious. Hernáez et al. [36] and Vialás et al. [35] used a proteomic strategy based on cell shaving of extracellular peptides to identify surface proteins in C. albicans yeast and hyphae forms, using Nano-LC followed by off-line MS/MS for peptide separation and identification. By this method, many novel surface proteins were identified that had not previously been reported as being on the cell surface. These proteins included some with unknown functions and aerobic respiration-related and ribosomal proteins, such as Rpl15A, Rps16A and Rps4A. In the present work, growing yeast and hypha cells were analyzed using the same strategy and more sensitive separation and identification equipment, enabling the identification of a larger number of proteins in each sample. In addition, a phenotypic analysis in vitro and/or in vivo of mutants with undescribed function of 17 identified proteins was performed to investigate their role in cell wall biogenesis, stress and virulence.
2. Materials and methods
2.1. Strains and growth conditions
C. albicans SC5314 [37] was used as wild type in this work. C. albicans mutant strains used in the in vitro and in vivo phenotypic studies were acquired from Noble collection [38] stored in the Fungal Genetics Stock Center (Kansas City, Missouri USA) [39]. C. albicans strains were maintained on YPD (1% yeast extract, 2% peptone, and 2% glucose) agar plates at 30 °C. Yeast cells were pre-cultured in liquid YPD medium (10 g/l yeast extract, 20 g/l peptone and 20 g/l glucose) in a rotary shaker at 200 rpm and 30 °C overnight. For the cell surface study, yeast cells were collected at an OD of 0.6 ± 0.2, which corresponds to logarithmic growth phase, and washed. A total of 5× 106 cells/mL were resuspended in Lee's medium [40] (0.2 g/l magnesium sulfate, 2.5 g/l dipotassium phosphate, 5 g/l sodium chloride, 5 g/l ammonium sulfate, 0.5 g/l L-alanine, 1.3 g/l Leucine, 1 g/l L-lysine, 0.1 g/l L-methionine, 0.07 g/l L-ornithine, 0.5 g/l L-phenylalanine, 0.5 g/l L-proline, 0.5 g/l L-threonine, 25 ml/l 50% glucose and 1 ml/l 0.1% biotin) at two different pHs, pH 4.2 for yeast growth and pH 6.7 for hyhae growth, and incubated at 37 °C for 6 h. At the end of the incubation period, cells were analyzed by light microscopy showing yeast shape at pH 4.2 and C. albicans hyphae at pH 6.7. To determine the generation time, strains were cultured in liquid YPD at 30 °C with OD600 0.1. Absorbance of the cultures was measured at 600 nm every hour. Linear regression was used to calculate specific growth rate and generation time. For phagocytosis and cytotoxicity assays, RAW 264.7 murine macrophages were cultured in RPMI 1640 medium supplemented with antibiotics (penicillin 100 U/ml and streptomycin 100 μg/ml), L-Glutamine (2 mM) and 10% heat-inactivated fetal bovine serum (FBS) at 37 °C in a humidified atmosphere containing 5% CO2. The FaDu oral epithelial cell line was obtained from the American Type Culture Collection and maintained in Eagle's minimum essential medium with Earle's balanced salt solution (Irvine Scientific) supplemented with 10% fetal bovine serum, 1 mM pyruvic acid 2 mM L-glutamine and 0.1 mM non essential amino acids.
2.2. Surface shaving
For surface protein identification, Hernáez et al. [36] method was used. Briefly, after 6 h of culture under different conditions, cells were collected and washed several times. Subsequently, cells were resuspended in 800 μl of 25 mM ammonium bicarbonate buffer (pH 8.0). A total amount of 5 μg of trypsin (Roche) in the presence of 0.1 mM DTT was added to the cell suspensions. After incubation at 37 °C for 5 min in a rotary shaker at 600 rpm, proteolytic reactions were stopped by adding 0.1% trifluoroacetic acid (TFA) v/v. Samples were centrifuged and supernatants were collected. The peptide supernatants were filtered through 0.22 μm pore-size filters (Millipore). Cell lysis was controlled by propidium iodide (PI) staining using flow cytometry in case of yeast and by fluorescence microscopy in case of hyphae. Before mass spectrometry analysis, the peptide supernatants were cleaned up with a microcolumn filled with Poros 50 R2 packing (PerSeptive Biosystems). Peptides were eluted with 80% acetonitrile (ACN) in 0.1% TFA, dried in a Speed-vac and resuspended in 0.1% formic acid. The samples were stored at −20 °C before nano-LC–MS/MS analysis.
2.3. LTQ-Orbitrap Velos analysis and protein identification
Prior to nano-LC–MS/MS analysis, all peptide samples were purified and desalted using C18-A1 ASY-column 2 cm pre-column (Thermo Scientific) and then eluted onto a Biosphere C18 column (Nano-Separations). Peptides were separated with a 140 min gradient (110min from 0 to 40% Buffer B; Buffer A: 0.1% formic acid/2% acetonitrile; Buffer B: 0.1% formic acid in acetonitrile) at a flow-rate of 250 nl/min on a nano-Easy HPLC (Proxeon) coupled to a nano-electrospray ion source (Proxeon). Mass spectra were acquired on the LTQ-Orbitrap Velos (Thermo Scientific) in the positive ion mode. Full-scan MS spectra (m/z 400/1400) were acquired in the Orbitrap with a target value of 1,000,000 at a resolution of 60,000 at m/z 400 and the 15 most intense ions were selected for collision induced dissociation (CID) fragmentation in the LTQ with a target value of 10,000 and normalized collision energy of 38%. Precursor ion charge state screening and monoisotopic precursor selection were enabled. Singly charged ions and unassigned charge states were rejected. Dynamic exclusion was enabled with a repeat count of 1 and exclusion duration of 30 s.
2.4. Protein identification and analysis
Protein identification from raw data was carried out using a licensed version of search engine MASCOT 2.3.0 with Proteome Discoverer software version 1.4.1.14 (Thermo Scientific). A database search was performed against the CGD21 database (6221 sequences). Search parameters were oxidized methionine as variable modification, peptide mass tolerance 10 ppm, 1 missed trypsin cleavage site and MS/MS fragment mass tolerance of 0.8 Da. In all protein identification, the FDR was b1%, using a Mascot Percolator [41], with a q-value of 0.01. As an estimation of the relative protein abundances the normalized spectral abundance factor (NSAF) was used [42], and the average of the normalized values was calculated. The MS output files have been submitted to PeptideAtlas through the PeptideAtlas Submission System (PASS) with identifier PASS00446.
2.5. Low Temperature Scanning Electron Microscopy (LTSEM)
Untreated C. albicans cells were mechanically fixed onto the specimen holder of a cryotransfer system (OxfordCT-1500), plunged into subcooled liquid nitrogen, and then transferred to the microscope's preparation unit via an air-lock transfer device. The frozen samples were cryofractured and etched for 2 min at −90 °C. After ice sublimation, etched surfaces were gold sputter coated and the specimens were then placed on the cold stage of the SEM chamber. Fractured surfaces were observed under a DSM960 Zeiss SEM microscope at −135 °C.
2.6. Assays to test susceptibility to stressors
C. albicans wild type and mutant strains were grown overnight in liquid YPD medium at 200 rpm and 30 °C. Yeast suspensions were adjusted at OD600 0.8 and serial 5 μl of 10-fold dilutions were spotted on YPD agar plates. For heat-shock stress assay the plates were incubated at 30 °C, 37 °C, 42 °C and 45 °C. For treatment with cell wall-, osmotic and oxidative-perturbing agents, the plates were supplemented with Calcofluor white (28 μg/ml), Congo red (175 μg/ml), sorbitol (1.5 M), KCl (1.2 M) and menadione (0.2 mM). Identical plates of Cell wall perturbing agents were prepared supplemented with 1 M Sorbitol. The wild-type and mutant strains were assayed for ability to withstand lethal dose of hydrogen peroxide. Strains were grown at 0.8 OD at 600 nm on 5 ml YPD liquid media containing hydrogen peroxide to a concentration of 100 mM incubated at 30 °C for 90 min. Five μl of each culture was spotted on YPD plates at different periods of time: 0, 2, 5, 10, 15, 30, 60 and 90 min. To assay the utilization of the polyol mannitol as a carbon source, strains were spotted onto mannitol agar plates (20 g/l mannitol, 5 g/l ammonium sulfate, 1.7 g/l nitrogen base and 2.2 g/l amino acids mix). After incubation at 30 °C for 24–48 h, the plates were imaged.
2.7. Filamentation assays
For invasive and filamentous growth on solid media, strains and wild-type were plated onto Spider agar (2% nutrient broth, 2% mannitol, 0.8% K2HPO4, 5.4% Bacto Agar) and YPD plates supplemented with 10% fetal bovine serum at a density of 30 CFUs/plate. Spider plates were incubated at 30 °C and 37 °C, and 10% fetal bovine serum plates at 30 °C for 7 days.
2.8. C. albicans’ phagocytosis assay and cytotoxicity measurement
For phagocytosis assay, macrophages were plated onto 18-mm glass sterile coverslips placed in 24-well plates. C. albicans strains were pre-labeled with 1 μM Oregon Green 488 (Molecular Probes) in the dark with gentle shaking (30 °C) for 1 h. Macrophages were confronted with the yeast at a MOI (multiplicity of infection; macrophage/yeast ratio) of 1 at 37 °C and 5% CO2. Interaction was stopped after 45 min, 1.5 and 3 h and cells were then washed with ice-cold PBS and fixed in 4% paraformaldehyde for 30 min. To distinguish between internalized and attached/non-ingested yeasts, C. albicans cells were counterstained with 2.5MCalcofluorwhite (Sigma) for 15 min in the dark. The number of ingested cells (green fluorescence) and/or adhered/non-ingested (Calcofluor white blue fluorescence) was quantified by fluorescence microscopy with FITC and UV [43]. Three different replicates with two different slides were prepared for each MOI and time point. At least 400 C. albicans cells were scored per slide, and results were expressed as the percentage of yeasts internalized by macrophages. For cytotoxicity measurement, macrophages were co-incubated (in a new complete media without phenol red (pH indicator) to avoid the background in the LDH test) with C. albicans strains at a MOI (multiplicity of infection; macrophage/bacteria ratio) of 1 during 3 and 8 h. Staurosporine 5 mM was used as a positive control. After the incubation, LDH was measured with the Cytotoxicity Detection KitPLUS (Roche) according to the manufacturer's protocol.
2.9. Endocytosis and damage assays
The number of organisms that were endocytosed by FaDu cells was determined using a differential fluorescence assay as described previously [44]. Briefly, 2.5 × 105 FaDu epithelial cells were grown on fibronectin-coated glass coverslips placed in a 24-well tissue culture plate and were infected with 105 cells of either C. albicans in RPMI 1640 medium the next day. After a 90 min incubation, the cells were rinsed once with PBS to remove the organisms that were not cell-associated and then fixed in 3% paraformaldehyde. The non-endocytosed organisms were stained for 1 h with an anti-C. albicans rabbit antiserum conjugated with Alexa 568 (Meridian Life Sciences, Inc.). After rinsing with PBS, the FaDu cells were permeabilized in 0.5% Triton X-100 for 10 min, and then the cell-associated organisms (defined as the endocytosed plus non endocytosed organisms) were stained with the anti-C. albicans rabbit serum conjugated with Alexa 488 (Molecular Probes). The coverslips were observed with epifluorescence microscope. The number of organisms’ endocyted by the FaDu cells was determined by subtracting the number of cell-associated organisms (labeled with Alexa 568, which fluoresced red) from the total number of organisms (labeled with Alexa 488, which fluoresced green). Organisms that were partially internalized were counted as being endocytosed. At least 100 organisms were counted on each coverslip, and all experiments were performed in triplicate.
The extent of damage caused by the various C. albicans strains to the FaDu cell line was measured using a 51Cr release assay as described previously [45]. The host cells were grown in a 96-well tissue culture plate containing detachable wells and incubated overnight with Na251CrO4 (MP Biomedicals, Inc., Irvine, CA) per well. The following day, the unincorporated tracer was aspirated and the cells were washed with HBSS and then infected with 105 organisms in RPMI 1640. To measure the spontaneous release of 51Cr, uninfected FaDu cells were exposed to medium alone. After 3 h incubation at 37 °C, the upper 50% of medium was removed from each well and then the wells were manually detached from one another. The amount of 51Cr in the aspirates and the well was determined by gamma counting. After correcting for the amount of 51Cr incorporated in each well, the specific release of 51Cr was calculated by the formula: (experimental release – spontaneous release)/(total incorporation – spontaneous release). Experimental release was the amount of 51Cr released into the medium by cells infected with C. albicans. Spontaneous release was the amount of 51Cr released into the medium by uninfected host cells. Total incorporation was the sum of the amount of 51Cr released into the medium and remaining in the host cells. Each experiment was performed in triplicate at least three different times.
2.10. Mouse model of oropharyngeal candidiasis
The virulence of the different C. albicans strains was assessed using the mouse model of oropharyngeal candidiasis as described previously [46]. Briefly, five male BALB/c mice per strain of C. albicans were immunosuppressed by subcutaneous injection with 225 mg/kg of cortisone acetate (Sigma-Aldrich) on days −1, 1 and 3 relative to the day of infection. On the day of infection, each mouse was anesthetized and inoculated sublingually for 75 min with a swabs saturated with 106 cells per ml. After 5 days of infection, each mouse was sacrificed, the tongue was excised, weighted, and homogenized, and the number of CFUs was determined. The animal experiments were approved by the Animal Care and Use Committee at the Los Angeles Biomedical Research Institute.
3. Results
3.1. Global analysis of C. albicans yeast and hypha-surface proteins by cell shaving
In this work, a modified and optimized proteomic strategy to identify surface proteins in C. albicans cells was followed [35,36]. Live cells were treated with trypsin to digest the cell surfaces. Cell lysis was monitored by flow cytometry and fluorescence microscopy in yeasts and hyphae, respectively, to control for cell lysis after trypsin digestion and avoid intracellular protein contamination. Aliquots of cells were taken from each sample before and after trypsin digestion and tested in each treatment. There was no significant increase in the percentage of PI population after incubation with the trypsin, being always under 0.7%. In addition, the cell surface was imaged by scanning electron microscopy after cryofracture before and after trypsinization. Before digestion, the cell surface was smooth, and it became wrinkled after digestion (Supplemental Fig. S1).
Proteomic analysis of yeast and hyphae was performed with proteins identified in at least two replicates with more than two peptides in each and proteins identified with one peptide in at least three replicates. A total of 438 and 928 proteins were identified in yeast and hypha samples, respectively (Supplemental Tables S1 and S2). The data corresponded to three biological replicates for yeast samples and four biological replicates for hyphal samples. They are deposited in the PeptideAtlas database [47], and information on each protein can be accessed through the CGD.
The identified proteins were classified in eight different categories based on their extracellular localization and their function related to cell wall maintenance, stress resistance or pathogenesis. These categories include GPI-anchored proteins, cell wall organization or biogenesis related proteins, cell surface proteins, proteins involved in pathogenesis or stress response, plasma membrane proteins, open reading frames (ORFs) and other identified proteins (Table 1). The Orfs are proteins that are unnamed in CGD and include proteins with verified feature type, meaning that there is experimental evidence for the existence of a gene product, or uncharacterized, without experimental evidence. The ‘other identified proteins’ category includes proteins with no relationship to the other categories above (Table 1). In both samples, this category included almost 40% of the proteins identified. Table 1 also includes the number of proteins in each type of sample and the corresponding percentage based on the total number of proteins in the sample. Proteins were classified according to Gene Ontology (GO) terms and the groups were ordered by importance in this work. This categorization is hierarchical and mutually exclusive, meaning that proteins included in one group are not classified again in any of the other groups.
Table 1.
Number and percentage distribution of different proteins identified by shaving C. albicans SC5314 yeast and hyphae cells.
| Categories | Yeast | Hypha | ||
|---|---|---|---|---|
| N° of proteins | % | N° of proteins | % | |
| GPI-anchored | 19 | 4.34 | 26 | 2.8 |
| Cell wall organization or biogenesis | 23 | 5.25 | 50 | 5.39 |
| Cell surface | 60 | 13.7 | 64 | 6.9 |
| Pathogenesis | 13 | 2.97 | 34 | 3.66 |
| Stress | 53 | 12.1 | 119 | 12.82 |
| Plasma membrane | 24 | 5.48 | 87 | 9.38 |
| Orf's | 53 | 12.1 | 172 | 18.53 |
| Other identified proteins | 193 | 44.06 | 376 | 40.52 |
| Total | 438 | 100 | 928 | 100 |
Proteins were classified in the 8 designated protein categories according to Candida Genome Database.
3.2. Comparative analysis of C. albicans yeast and hyphae-surface proteins
Of the proteins identified, hyphae had slightly more GPI proteins than did yeast (Table 1). The number of cell surface proteins is similar in the two different samples, however looking at the percentage based on the total proteins identified per sample this group is more highly enriched in yeast (13.7%) than in hyphae (6.9%). In the cell wall, pathogenesis and stress response-related identified proteins twice asmany of these proteins were identified in hyphae as compared to yeast, while the percentage of proteins in these categories was similar for both morphologies. Moreover, three-fold more proteins in the plasma membrane and Orfs categories were identified in hypha. In addition, the averaged normalized spectral abundance factor (NSAF) [42] of each sample was performed to analyze the relative abundance of the proteins. It was noticeable that in the top 20 proteins of both samples, 14 were present in both yeast and hyphae, with six cell surface proteins (Tdh3, Adh1, Eno1, Pgk1, Pdc11 and Tal1), three stress response-related (Ahp1, Hsp12 and Orf19.4216), one related to pathogenesis (Wh11) and four categorized as other (Rpl38, Rps14b, Rps20 and Tef2) (Table 2). In the case of yeast, cell surface proteins were more abundant (seven) followed by other surface proteins (six), stress-related (four), Orfs (two) and pathogenesis-related (one). The distribution of hyphal was slightly different, with other surface proteins being the most representative with nine proteins, followed by cell surface (seven), stress-related (three) and pathogenesis-related proteins (one).
Table 2.
Ranking of the top twenty proteins according to the averaged normalized spectral abundance factor (NSAF) in yeast and hyphal C. albicans cells.
| Yeast | Hyphae | ||
|---|---|---|---|
| Protein | NSAF | Protein | NSAF |
| Tdh3 | 0.0230 | Tdh3 | 0.0238 |
| Wh11 | 0.0213 | Adh1 | 0.0089 |
| Hsp12 | 0.0171 | Eno1 | 0.0087 |
| Orf19.4216 | 0.0170 | Pgk1 | 0.0084 |
| Pgk1 | 0.0127 | Wh11 | 0.0083 |
| Eno1 | 0.0113 | Pdc11 | 0.0082 |
| Rps14b | 0.0102 | Rpl38 | 0.0078 |
| Crd2 | 0.0098 | Rps14b | 0.0075 |
| Mp65 | 0.0094 | Tef2 | 0.0073 |
| Cyp1 | 0.0089 | Orf19.4216 | 0.0070 |
| Orf19.3690.2 | 0.0087 | Hsp12 | 0.0070 |
| Pdc11 | 0.0087 | Tal1 | 0.0066 |
| Tal1 | 0.0083 | Ahp1 | 0.0065 |
| Glx3 | 0.0082 | Rps1 | 0.0064 |
| Tef2 | 0.0074 | Rpl14 | 0.0063 |
| Orf19.6415.1 | 0.0073 | Rps20 | 0.0062 |
| Rpl38 | 0.0073 | Rps3 | 0.0061 |
| Rps20 | 0.0072 | Ece1 | 0.0061 |
| Ahp1 | 0.0071 | Rpl23a | 0.0060 |
| Adh1 | 0.0070 | Rps13 | 0.0059 |
In grey, proteins that appear in both hits.
Venny analysis was carried out to find proteins common to both morphologies and proteins identified exclusively in a cellular morphology. A total of 943 unique proteins were detected. Of these, 423 proteins were present in both yeast and hyphae. The other 520 remaining proteins were found exclusively in yeast (15 proteins) and hyphae (505 proteins). All of these proteins were re-ordered in the eight groups detailed above and according to the morphology in which they were identified (common or exclusive to one of the morphologies) (Table 3). Again, the results showed that a large proportion of the proteins identified were present in both morphologies. The largest differences were found in cell wall-, pathogenesis-, stress response- and plasma membrane-related proteins, and clearly in the Orf's group, where more than twice as many proteins were identified in hyphae. In the GPI-anchor protein group, the hypha-specific protein Hyr1 was identified exclusively in this morphology. Sap10 and Als2 were yeast exclusive GPI proteins. In the case of cell wall-related proteins, the difference was higher, Xog1 was detected only in yeast, 28 proteins were detected only in hyphae and 22 were present in both. Clearly, more proteins are expressed on the surface of hyphae than on yeast. It is interesting to note that a high number of Orf's were detected in both samples, with 172 proteins identified in hyphae (Table 1), and with 53 in yeast (Table 3).
Table 3.
Categorization of proteins identified in C. albicans SC5314 surfome of yeast and hyphae.
| Categories | Common in Yeast and Hypha | Only identified in Yeast | Only identified in Hypha | Total |
|---|---|---|---|---|
| GPI-anchored | 17 | 2 | 9 | 28 |
| Als1, Cht2, Crh11, Ecm33, Ihd1, Pga4, Phr1, Phr2, Pir1, Plb4.5, Rbt1, Rbt5, Rhd3, Sap9, Ssr1, Utr2, Ywp1. | Als2, Sap10. | Als3, Eap1, Hyr1, Pga10, Pga45, Pga52, Pga53, Plb3, Sod5. | ||
| Cell wall organization or biogenesis | 22 | 1 | 28 | 51 |
| Act1, Agm1, Bgl2, Bmh1, Cmk2, Eng1, Gda1, Gfa1, Mnt1, Msb2, Pmi1, Ras1, Rho1, Sim1, Slk19, Sod1, Srb1, Sun41, Sur7, Tsa1, Yps7, Ypt31. | Xog1. | Arp2, Bud7, Cdc10, Chs5, Clc1, Cwh43, Dck1, End3, Flc2, Gsc1, Hog1, Kex2, Kre9, Lmo1, Mnn26, Pbs2, Pho85, Pmr1, Pmt1, Pmt2, Pmt4. Pop2, Rac1, Rvs161, Rvs167, Smi1, Snf1, Tps2. | ||
| Cell surface | 58 | 2 | 6 | 66 |
| Adh1, Atp1, Atp2, Cdc19, Cdc48, Cef3, Cht3, Coi1, Csp37, Ddr48, Efb1, Eft2, Egd2, Eno1, Fba1, Gpd2, Gph1, Gpm1, Hem13, Hsp104, Hsp21, Hsp70, Hsp90, Ino1, Kar2, Met6, Mp65, Orf19.2478.1, Pdc11, Pdi1, Pgk1, Pma1, Rbe1, Rpl10, Rpl13, Rpl14, Rpl17b, Rpl19a, Rpl20b, Rpl3, Rpl4b, Rpl6, Rps10, Rps6a, Rps7a, Rsp8a, Sam2, Scw11, Ssa2, Ssb1, Ssc1, Ssz1, Tal1, Tdh3, Tkl1, Tos1, Tpi1, Ugp1. | Fet99, Pho113. | Aip2, Cdr1, Fre10, Rax2, Rbt4, Sdh2. | ||
| Pathogenesis | 12 | 1 | 22 | 35 |
| Ade5.7, Age3, Asc1, Cdc42, Fas2, Msi3, Tfp1, Ttr1, Tup1, Vma2, Wh11, Yhb1. | Hex1. | Alo1, Cat1, Cdc11, Cla4, Crk1, Csh3, Gna1, Gpa2, Het1, Mlt1, Mts1, Nmt1, Not5, Obpa, Ole1, Rsr1, Slr1, Srv2, Ssd1, Ssn6, Vps27, Ypt72. | ||
| Stress Response | 51 | 2 | 68 | 121 |
| Ade1, Ade12, Ade13, Aha1, Ahp1, Atc1, Bcy1, Ccp1, Cdc54, Ded1, Dot5, Erf1, Erg13, Gis2, Glc7, Gln1, Glx3, Gnd1, Gre3, Hsp12, Imh3, Lsp1, Orf19.1340, Orf19.4216, Orf19.4246, Orf19.4622, Orf19.5281, Orf19.5620, Orf19.5943.1, Orf19.6358, Pet9, Pil1, Pol30, Prx1, Rdi1, Rhr2, Rpg1a, Rpl16a, Rpt4, Sbp1, Sgt2, Skp1, Smt3, Sub2, Sui2, Tma19, Trr1, Trx1, Ynk1, Ypt1, Zpr1. | Orf19.4150, Orf19.7196. | Apm1, Arf2, Bdf1, Bre1, Cct8, Cdc28, Cdc37, Cip1, Crm1, Cta3, Dhh1, Doa1, Erg1, Gea2, Gpd1, Grx3, Hat2, Hbr1, Hrr25, Hsp60, Hta1, Hta2, Lag1, Mcr1, Mxr1, Myo2, Ncb2, Orf19.2265, Orf19.2304, Orf19.239, Orf19.512, Orf19.5833, Orf19.5917.3, Orf19.6250, Orf19.6424, Orf19.7160, Orf19.86, Orf19.933, Osm2, Pob3, Ptc2, Pyc2, Rad23, Rad6, Rfa2, Rfc1, Rgd1, Rli1, Rpo21, Rp6, Sec18, Sec7, She3, Shp1, Sin3, Sis1, Spt5, Spt6, Svf1, Taf14, Tfg1, Tif34, Ura7, Vac8, Yck2, Yim1, Ypt52, Zrt2. | ||
| Plasma membrane | 23 | 1 | 64 | 88 |
| Ade17, Adh2, Cof1, Erg6, Erv25, Faa4, Glk1, Hgt6, Hgt7, Ist2, Mdg1, Met15, Orf19.1564, Orf19.6160, Orf19.6553, Pfy1, Pgi1, Pho88, Sac6, Tif, Tom22, Tom70, Ycp4. | Fet3. | Ali1, Apm4, Atp17, Atp3, Atp4, Atp7, Cbr1, Chc1, Cox13, Cox4, Cox5, Cox6, Cox9, Ctr1, Cyt1, Ddi1, Dpm1, Elf1, Emp24, Erg11, Frp3, Gap4, Gca1, Gut2, Hmg1, Hom2, Kin2, Lem3, Mir1, Nde1, Orf19.1840, Orf19.1970, Orf19.3003, Orf19.3235, Orf19.3290, Orf19.3335, Orf19.3430, Orf19.409, Orf19.4396, Orf19.4864, Orf19.5006.1, Orf19.5095, Orf19.5340, Orf19.5669, Orf19.6082, Orf19.7310, Pan1, Phm7, Por1, Pst3, Qcr2, Rho3, Rip1, Sec26, Sec4, Sec61, Sec62, Shm2, Stt3, Tcp1, Tim23, Tim50, Tom20, Vma5. | ||
| Orf's | 50 | 3 | 122 | 175 |
| Other identified proteins | 190 | 3 | 186 | 379 |
| Total | 423 | 15 | 505 | 943 |
Proteins were classified in the 8 designated categories according to Candida Genome Database (CGD) Gene Ontology (GO) terms (in parentheses the GO identifier): Cell wall organization or biogenesis (71554), cell surface (9986), pathogenesis (9405), response to stress (6950) and plasma membrane (5886). GPI-anchored category is based on Richard et al. [2]. Orf's category refers to proteins without standard name in CGD. Other identified proteins category is composed by proteins not classified in previous sections.
3.3. Phenotypic and virulence analysis of interesting identified proteins
Among the identified proteins, 17 were selected to investigate their possible involvement in the formation of the cell wall as the outermost structure of the cell, due to their being proteins of unknown function, their implication in the interaction with the host and the fact that they have been characterized only in large-scale studies. The availability of the mutants and the morphologic form of the organism in which the protein had been identified were also used as criteria to select the proteins. These 17 proteins are listed in Table 4 and they are classified in the following categories: GPI (two), cell surface (one), stress related (one), plasma membrane (six) and Orf's (five). Two proteins classified in the ‘Other identified proteins’ category were selected to be studied, Mci4 and Ptp3. The first was selected because its feature type at CGD is uncharacterized and the second because it is more abundant in hypha.
Table 4.
List of C. albicans selected proteins for the phenotypic and virulence analysis of the corresponding mutants.
| CGD Accesion | Protein name | Cathegory annotated (CGD) | Descriptiona | Replicates with protein (peptides for each replicate) | |
|---|---|---|---|---|---|
| Yeast | Hyphae | ||||
| orf19.1690 | Tos1 | Cell surface | Protein similar to alpha agglutinin anchor subunit | 3 (11,9,11) | 4 (10,10,10,8) |
| orf19.1710 | Ali1 | Plasma membrane | Putative NADH-ubiquinone oxidoreductase | - | 2 (2,2) |
| orf19.2451 | Pga45 | GPI | Putative GPI-anchored cell wall protein | - | 4 (4,5,5,2) |
| orf19.2570 | Mci4 | Other | Putative NADH-ubiquinone dehydrogenase | - | 3 (1,1,1) |
| orf19.287 | - | Orf's | Putative NADH-ubiquinone oxidoreductase subunit | - | 3 (1,1,1) |
| orf19.3060 | - | Orf's | OPutativedolichyl-diphoshooligosaccharide-proteinglycotransferase | - | 3 (3,3,2) |
| orf19.3290 | - | Plasma membrane | Plasma membrane-localized protein | - | 3 (2,2,1) |
| orf19.3335 | - | Plasma membrane | Plasma membrane protein of unknown function | - | 4 (1,1,4,4) |
| orf19.5285 | Pst3 | Plasma membrane | Putative flavodoxin | - | 4 (2,3,3,8) |
| orf19.5286 | Ycp4 | Plasma membrane | Putative flavodoxin | 3 (1,1,2) | 4 (2,2,3,2) |
| orf19.5352 | - | Orf's | Protein with a predicted magnesium transporter domain | - | 3 (1,1,1) |
| orf19.5760 | Ihd1 | GPI | GPI-anchored protein | 3 (2,2,1) | 4 (2,2,2,2) |
| orf19.6553 | - | Plasma membrane | Membrane-localized protein of unknown function | 2 (3,2) | 4 (1,1,1,1) |
| orf19.7196 | - | Stress | Putative vacuolar protease | 2 (2,2) | - |
| orf19.7328 | - | Orf's | Protein with a staphylococcal nuclease domain | - | 4 (14,8,8,7) |
| orf19.7590 | - | Orf's | Putative NADH-ubiquinone oxidoreductase | - | 3 (2,5,4) |
| orf19.7610 | Ptp3 | Other | Putative protein tyrosine phosphatase | - | 4 (3,3,6,7) |
Description according to Candida Genome Database (CGD)
With this purpose, the homozygotic mutants were acquired from the Fungal Genetics Stock Center (FGSC) and different phenotypic assays were carried out. Strain SC5314 was used for the study of surfome. Although the wild type of the mutant collection is the SN250, we validated that both strains have the same behavior under all conditions tested, thus SC5314 can be used as a reference strain.
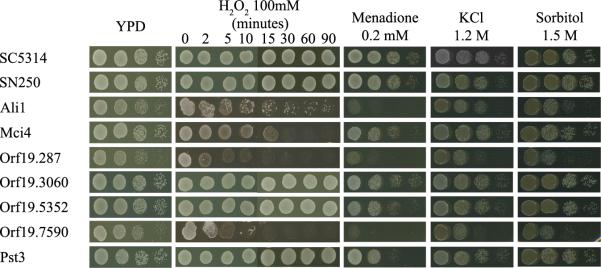
The assay conditions were separated into the broad categories of stress (including temperature, cell wall perturbing agents, osmotic and oxidative stress), morphology and nutrition. The sensitivity of the mutants to 175 μg/ml Congo red and 28 μg/ml Calcofluor white, compounds that interfere in the cross-linking of cell wall components [9,48], was tested. Several mutants were shown to be sensitive to those compounds, with ali1Δ, orf19.287Δ, orf19.3060Δ, orf19.5352Δ and pst3Δ sensitive to both Calcofluor white and Congo red, and orf19.7590Δ and tos1Δ slightly sensitive to Calcofluor white (Fig. 1 and Table 5). Sensitivity to these compounds was suppressed on osmotic medium stabilized with sorbitol 1M, suggesting that the sensitivity observed was due to a defect in the cell wall [49]. Heat shock stress was tested growing the strains at different temperatures. All strains grew well at 37 °C, while at 45 °C ali1Δ, orf19.287Δ, orf19.3060Δ, orf19.3290Δ, orf19.7590Δ and pst3Δ had an altered growth (Table 5).
Figure 1. Phenotypic analysis of C. albicans mutants to cell wall perturbing agents.
Sensitivity to Calcofluor white and Congo red of the wild-type strains SC5314 and SN250 and C. albicans mutants.
Table 5.
Phenotypes of C. albicans mutants.
| C. albicans strain | Generation time 30 °C (min) | Stress | Morphology | Nutrition | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cell Wall | Osmotic | Oxidative | T° | |||||||||||
| Sensitivity to Calcofluor White 28 μg/ml | Calcofluor White 28 μg/ml + 1 M sorbitol | Sensitivity to Congo Red 175 μg/ml | Congo Red 175 μg/ml + 1 M sorbitol | Sensitivity to Sorbitol 1.5 M | Sensitivity to KCl 1.2 M | Sensitivity to Menadione 0.2 mM | Sensitivity to H2O2 100 mM | 45 °C | Serum 10% 37 °C | Spider 30 °C | Spider 37 °C | C-Source: Mannitol | ||
| SC5314 | 72 | Control Strain (wild-type) | ||||||||||||
| SN250 | n.d. | Collection Control Strain | ||||||||||||
| ali1 | 100 | + | √ | + | √ | + | + | + | + | + | + | + | + | + |
| mci4 | 88 | + | + | + | + | + | + | + | ||||||
| orf19.287 | 96 | + | √ | + | √ | + | + | + | + | + | + | + | + | |
| orf19.3060 | n.d. | + | √ | + | √ | + | ||||||||
| orf19.3290 | n.d. | + | + | + | ||||||||||
| orf19.5352 | 71 | + | √ | + | √ | |||||||||
| orf19.7590 | 99 | + | √ | + | + | + | + | + | + | + | ||||
| pst3 | n.d. | + | √ | + | √ | + | + | |||||||
| tos1 | 73 | + | √ | |||||||||||
Sensitivity to the compounds was indicated with “+” and means reduction of growth relative to the controls SC5314 and SN250. Blank box stand for the same growth as the controls. “n.d.” or Not Determined. Stress media growth, colony morphology and nutrient media growth phenotypes were indicated in the table. The data shown are from at least three replicates. 8 out 17 mutants studied (ihd1, orf19.3335, orf19.6553, orf19.7196, orf19.7238, pga45, ptp3 and ycp4) not exhibit any phenotype.
Oxidative stress was tested using 0.2 mM menadione, which can generate reactive oxygen species (ROS) by redox cycling which leads to stress, and 100 mM H2O2, a ROS that damages cellular components, as described in Section 2. Three mutants were sensitive to both H2O2 and menadione, ali1Δ, orf19.287Δ and the orf19.7590Δ. The pst3Δ mutant was sensitive just to menadione and mci4Δ and ycp4Δ just to H2O2 (Fig. 2 and Table 5). Strains were also exposed to osmotic stress by adding 1.5 M sorbitol and 1.2 M KCl to the medium. The ali1Δ, orf19.3290Δ and mci4Δ mutants were sensitive to both compounds while the orf19.287Δ mutant was sensitive only to KCl (Fig. 2 and Table 5).
Figure 2. Oxidative and osmotic-perturbing agents assays in C. albicans mutants.
Mutants were exposed to oxidative-perturbing agents (H2O2 and menadione) and osmotic-perturbing agents (KCl and sorbitol) onto YPD plates containing the indicated amounts of the compound to be tested.
In addition, the capacity of the mutants to form filaments was studied. When growing on YPD agar supplemented with 10% serum the ali1Δ, orf19.7590Δ, orf19.287Δ and mci4Δ mutants had a completely smooth colony morphology showing a severe defect in the yeast to hypha transition (Fig. 3A and Table 5). On Spider agar, the same mutants grew very poorly and very small, barely visible colonies appeared (Fig. 3B). This phenotype could be due to inability to metabolize the mannitol carbon source in Spider medium. For this reason, growth in mannitol agar plates was assayed. As expected, the mutant strains exhibit impaired growth on this medium, showing that they had an impaired ability to use mannitol as a carbon source (Fig. 3C and Table 5). In addition to investigating the growth of these mutants under different stress conditions, we also tested their growth at 30 °C to determine if any had a global growth defect. All strains had approximately the same growth rate to the parental strain, with the exception of ali1Δ, orf19.7590Δ, orf19.287Δ and mci4Δ mutants, which grew more slowly than the wild-type strain (Table 5).
Figure 3. Yeast-to-hypha transition assays in C. albicans mutants.
(A) Yeast-tohypha transition in YPD plates supplemented with 10% serum incubated at 37 °C for 7 days. (B) Yeast-to-hypha transition in Spider plates incubated at 37 °C for 7 days. (C) C. albicans mutants with growth defect on Spider plates exhibited a reduced growth rate on the C-source: mannitol medium.
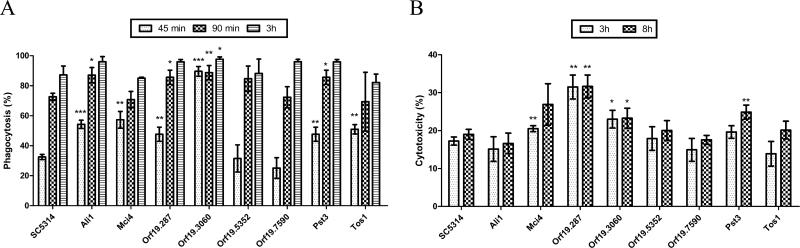
Based on these results, the subsequent analyses were focused on the following eight mutants: ali1Δ, mci4Δ, orf19.287Δ, orf19.7590Δ, pst3Δ, tos1Δ, orf19.3060Δ and orf19.5352Δ. These mutants were sensitive to cell wall disturbing agents, oxidative stress and osmotic stress, and in the case of ali1Δ, mci4Δ, orf19.287Δ and orf19.7590Δ had filamentation defects and were unable to use mannitol as a carbon-source (Table 5). The interaction of the eight C. albicans mutants and the parental C. albicans SC5314with RAW264.7 macrophages was studied. Phagocytosis by RAW264.7 macrophages were evaluated at an MOI of 1:1 and at different interaction times. As shown in Fig. 4A, at earlier time points, the ali1Δ, mci4Δ, orf19.287Δ, pst3Δ, tos1Δ and orf19.3060Δ mutants were phagocytosed statistically significant more than the wild-type strain. This difference was no longer evident at the 3 h time point for all strains tested.
Figure 4. Murine macrophage response to C. albicans mutants.
(A) Quantification of phagocytosis of C. albicans yeasts at 45 min, 1.5 and 3 h of coincubation. (B) Cytotoxicity of the different mutants of C. albicans in RAW 264.7 macrophages at 3 and 8 h of coincubation. Data are represented as mean ± SD (n=3), and statistical significance relative to wild-type is indicated (*, p<0.05; **, p<0.01; ***, p<0.001).
To determine the capacity of the different C. albicans mutants to damage RAW 264.7 macrophages, the lactate dehydrogenase (LDH) cytotoxicity detection kit was used to measure the amount of LDH released from the damaged cells into the medium. As shown in Fig. 4B, C. albicans mci4Δ, orf19.287Δ, pst3Δ and orf19.3060Δ caused significantly more damage to the macrophages as compared to the wild-type strain at one or both time points.
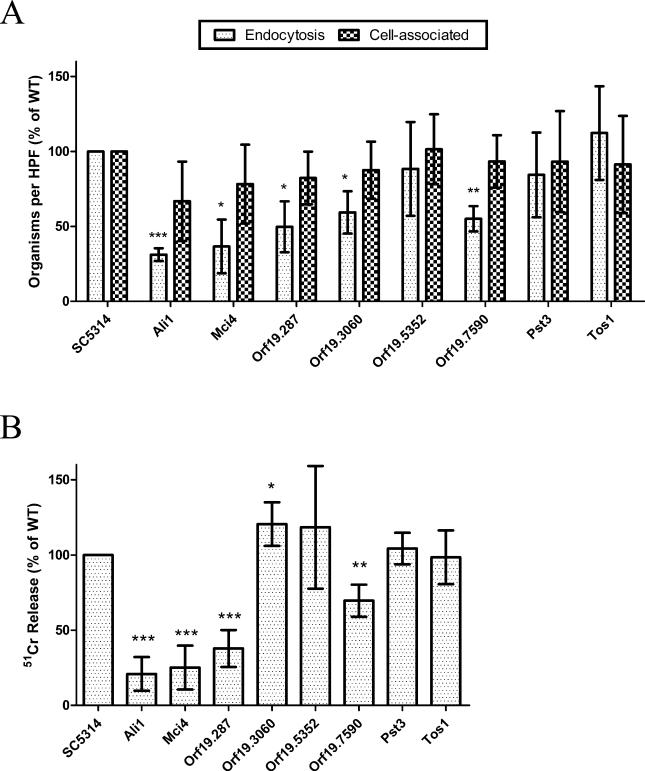
Fungal invasion of the superficial epithelial lining of the oral mucosa is an important feature of oropharyngeal candidiasis [50]. For this reason, the interactions of these mutants with the FaDu oral epithelial cell line in vitro were tested. After 90 min of infection, the number of cell-associated (adherent) organisms was similar to that of the wild type strain for all the mutants (Fig. 5A). At this time point, the endocytosis of the ali1Δ, mci4Δ, orf19.287Δ, orf19.3060Δ and orf19.7590Δ mutants was significantly less than that of the wild-type strain (Fig. 5A). Of note, the ali1Δ, mci4Δ and orf19.287Δ mutants germinated poorly on the FaDu cells and grew mainly as yeast, which likely explains their impaired endocytosis.
Figure 5. Role of the proteins for the adherence, invasion and induce damage to FaDu oral epithelial cells.
(A) The indicated strains of C. albicans were incubated with the FaDu oral epithelial cell line for 90 min, after which the numbers of endocytosed and cell-associated organisms were determined by a differential fluorescence assay. (B) The indicated host cells were loaded with 51Cr and then incubated with the indicated strains of C. albicans for 3 h. The extent of host cell damage was measured by the release of 51Cr into the medium. Results are the mean standard deviation of three experiments, each performed in triplicate. Data are represented as mean ± SD (n=3), and statistical significance relative to wild-type is indicated (*, p<0.05; **, p<0.01; ***, p<0.001).
Next, the extent of damage caused by the different C. albicans mutants to FaDu oral epithelial cells was investigated. The ali1Δ, mci4Δ, orf19.287Δ and orf19.7590Δ mutants caused significantly less damage to the cells compared with the wild-type strain (Fig. 5B). Interestingly, the orf19.3060Δ mutant caused slightly increased epithelial cell damage as compared with the wild-type strain.
The diverse phenotypes shown in the mutant strains’ in vitro test suggest that these mutants might have increased virulence during oropharyngeal infection. This hypothesis was tested using a mouse model of OPC. The oral fungal burden of mice infected with either pst3Δ, tos1Δ, orf19.3060Δ or orf19.5352Δ strains was similar to that of mice infected with the wild-type strain (Supplemental Fig. S2). Therefore, these proteins do not appear to be involved in virulence during oropharyngeal candidiasis.
4. Discussion
4.1. Cell shaving for the identification of C. albicans yeast and hyphae surface protein profile
This work is mainly based on the comprehensive study of the surface proteome of C. albicans and the selection of some identified and unknown-function proteins to analyze their relevance in the cell surface, stress and virulence. For the identification of surface proteins of both yeast and hyphae, a proteomic approach based on cell surface shaving through live cell trypsin digestion and identification by MS was used. A total of 438 and 928 proteins were identified in yeast and hyphae morphology, respectively (Supplemental Tables S1 and S2). Other previously published proteomic analysis of C. albicans surface in yeast and/or hyphal morphology reported a smaller number of detected proteins [31–36]. Even using the same methodology optimized to identify surface proteins in C. albicans yeast cells by Hernaez et al. [36] and by Vialás et al. [35], the number of proteins identified in this work is much higher. Thus the proteomics approximation used has been validated and improved in this work by using LTQ-Orbitrap Velos, an ultra-high resolution mass analyzer with increased sensitivity, versus the MALDI TOF/TOF used in previous works. Moreover, also the proportion of proteins identified in each of the more interesting categories is more enriched. Among the new identified proteins Als2, Cht2, Crh11, Ihd1, Pir1 and Sap9-10 were found. All of these were described as GPI-anchored proteins by Richard and Plaine [4].
Several of the identified proteins are classical metabolic cytoplasmic proteins, such as Eno1 or Tdh3. They have been described at the C. albicans and Saccharomyces cerevisiae cell wall using different proteomic approaches [35,36,51–54]. In all cell surface protein studies of different microorganisms using the shaving approach, this type of proteins was detected (reviewed in [55]). The mechanisms used for this type of proteins to reach the cell wall are under study and different possibilities have been proposed: non classical exporting/secretory mechanisms, membrane-vesicle structures in which proteins are trapped and residual cell lysis [55]. A recent work describes that C. albicans extracellular vesicles contains several classical cytoplasmic and membrane proteins, such as Eno1, Gpm1, Met6, Pdc11, Pgk1, Ssa2 or Tdh3 [18] suggesting that they could pass through the cell wall inside these vesicles. In the present work, more classical cytoplasmic proteins or proteins described as located at intracellular membranes were detected. This might be related with the more sensitivity of this study, but also, although cell lysis controls were carried out, a very low contamination of intracellular proteins cannot be absolutely discarded.
Regarding the different profiles of C. albicans cell surface proteins of yeast or hyphae, Heilmann et al. [33] proposed Rhd3, Sod4 and Ywp1 as indicative of yeast cells while in this work Rdh3 and Ywp1 were identified in both morphologies as in Sosinska et al. [34]. However, Als2 and Sap10 were detected only in yeast. For proteins exclusively detected in hyphae, some are adhesins and lytic enzymes associated with virulence and hyphal growth, such as Als3, Hyr1 or Sod5 [56–60] (Table 3). The detection of some of these proteins only in one of the morphological forms might suggest that they have a role in this morphology, but other factors can also regulate their expression; for example the pH could influence the composition of these proteomes. It is known that environmental pH has effects on C. albicans morphology and its ability to respond to stress [61]. In addition, proteins have a pH optimum for activity, thus their functionality depends on the media conditions, including secreted and surface exposed proteins. For example, the homologous PHR1 and PHR2 genes encode pH regulated cell surface glycosidases. Although Phr2 mRNA and protein levels are higher preferentially at acidic pH and Phr1 at neutral [34,62,63], here both proteins were identified in the two morphologies obtained at different pHs (Table 3). Consistent with our results, recent works, in which an enhanced mass analyzer with increased sensitivity was used, have described the increase in abundance of both proteins under wall stress and different pHs, strongly at pH 7.4, in the cell wall [64,65]. Furthermore, the expression of these proteins together with others such as Als1or Ihd1, is enhanced, but not exclusive, at determined pHs [66,67].
It is important to highlight the large number of Orf's identified (annotated as verified or uncharacterized at CGD), which totalled 175; the vast majority identified just in hyphae. Due to their implication in cell is unknown and based on published data and mutant availability, 17 mutants were selected to examine their involvement in different cellular roles and virulence.
4.2. New identified C. albicans proteins relevant for dimorphic transition, cell wall maintenance, oxidative or osmotic stress responses
The response of the selected mutants to different stresses, including temperature, oxidative, osmotic or cell wall stress, was analyzed. Half of the mutants did not show any sensitivity to the compounds or temperatures studied (eight out of 17, ihd1Δ, orf19.3335Δ, orf19.6553Δ, orf19.7196Δ, orf19.7238Δ, pga45Δ, ptp3Δ and ycp4Δ), meaning that these proteins do not have a relevant cell role in responding to these stresses. The other nine mutant strains, ali1Δ, mci4Δ, orf19.287Δ, orf19.3060Δ, orf19.3290Δ, orf19.5352Δ, orf19.7590Δ, pst3Δ and tos1Δ, presented sensitivity to some of the stresses tested.
In addition, ali1Δ, mci4Δ, orf19.287Δ and orf19.7590Δ showed filamentation defects in 10% serum agar plates and growing defects in the Spider plates containing mannitol. Indeed, these mutants were unable to grow using this C-source. These four proteins are described in CGD as putative NADH-ubiquinone oxidoreductases (Ali1, Orf19.287 and Orf19.7590) and a putative NADH-ubiquinone dehydrogenase (Mci4), and no S. cerevisiae orthologs are described. Previous work showed that C. albicans uses mannitol only after the exhaustion of glucose, increasing the activity of NAD-linked mannitol dehydrogenase [68]. A possible relation between this activity and the four NADHubiquinone-related proteins remains to be investigated. In fact, little is known about the mannitol catabolic process: just one protein has been directly related to it, Zcf7, a predicted Zn(II)2Cys6 transcription factor, to date [69]. Thus, it can only be suggested that these four proteins appear to be linked to the use of mannitol and to link specific cues in environment to colony phenotype. In addition, these strains showed sensitivities to the vast majority of stresses studied.
Out of the 17 initial mutants, eight had a marked increase in susceptibility to a diverse panel of stressors, correspond to eight proteins identified in hyphae, including the four NADH-ubiquinone related proteins noted before (Ali1, Mci4, Orf19.287 and Orf19.7590), Orf19.3060, Orf19.5352 and Pst3, and Tos1 that were detected in both morphologies. The Tos1 mutant, tos1Δ, together with ali1Δ, orf19.287Δ, orf19.3060Δ, orf19.5352Δ, orf19.7590Δ and pst3Δ, presented sensitive to cell wall-disturbing agents and this sensitivity was suppressed in osmotic medium stabilized with sorbitol 1 M, suggesting that the sensitivity observed was due to a defect in the cell wall [49]. Pst3 protein has an ortholog in S. cerevisiae, Pst2, described in the plasma membrane by electronic annotation in CGD, and curiously it was also identified in the DIGE proteomic analysis of C. albicans yeast-to-hypha transition, that analyzed mainly soluble proteins, but in this case Pst3 was detected as less abundant in hypha than in yeast cells [70]. S. cerevisiae Pst2 was also identified in a proteomic approach for the study of cell wall biogenesis, in accordance with the sensitivity shown for pst3Δ to the cell wall-disturbing agents [71] and is similar to a family of flavodoxin-like proteins which is in agreement with the increased oxidative stress sensitivity of pst3Δ mutant strain. It is important to note that orf19.3060Δ, orf19.5352Δ, and tos1Δ mutant strains only presented defects when they grew in the presence of the cell wall-disturbing agents; thus, they seem to have a cell wall-specific defect. The Orf19.5352 is an uncharacterized protein with a predicted magnesium transporter domain located in the membrane and no S. cerevisiae ortholog is described [72]; and therefore, it is difficult to correlate this predicted function with the cell wall damage. On the contrary, the orf19.3060Δ is defective in a protein with a possible role in protein N-linked glycosylation according to the function of its ortholog in S. cerevisiae, Swp1, a 30 kDa type I transmembrane [73]; which would lead to a weaker cell wall according to our results. Tos1 is a protein similar to an alpha-agglutinin anchor subunit. It was previously detected in extracellular vesicles and proteins secreted by C. albicans in different studies, but its function is not known [18,74–76] and according to our results would be involved in cell wall maintenance.
Of the other 10 proteins, where the mutant strains did not present any sensitivity to the stressors tested, or only to osmotic stress such as orf19.3290Δ, little is known about their function to date. The IHD1 gene was defined as part of the core filamentation network although Ihd1 is not essential for hyphal development according to current work [77].
4.3. C. albicans mutant strains produce different degrees of cell damage in host–pathogen interaction assays
Phagocytosis analysis reveals that six of the eight mutants used in this study showed an increase in engulfment by this type of immune cell. The mutation of these proteins could affect the composition of specific components of the C. albicans cell wall, leading to an increase in recognition by macrophages, as occurs with different O-linked and N-linked mannan-deficient strains in which the recognition by macrophages is increased [73]. Of these more phagocytized mutants, unexpectedly, the mci4Δ, orf19.287Δ, pst3Δ and orf19.3060Δ mutants, showed an increase in their ability to cause damage in the host cells. The increase in the amount of phagocytized yeast is crucial for macrophages in order to contain and kill Candida. The clearance of the internalized cells seems to be less efficient at increasing cytotoxicity to the macrophages, as occurs in macrophages co-incubated with C. albicans at different MOIs where the higher MOIs are more cytotoxic to the macrophages [78]. The orf19.3060Δ mutant showed the most important differences in phagocytosis. As commented above, this mutant strain is defective in a protein with a role in protein N-linked glycosylation. McKenzie et al. [73] assessed the contribution of C. albicans cell wall glycosylation in macrophage response and related the lack of MSN1 (N-glycosylation defect) with an increase in macrophage recognition and phagocytosis according to our data. The higher rate of orf19.3060Δ cells phagocyted and their normal hyphal formation can explain the increase in macrophage damage. The most cytotoxic strain was orf19.287Δ, which was more than 50% more cytotoxic than the wild type. Although this mutant strain is deficient in yeast-hypha transition, the large number of phagocyted cells could cause the increased damage by another mechanism, such as high secretion of proteins or enzymes involved in virulence. It is known that C. albicans is able to induce an apoptotic signal in macrophages during the interaction in the ratio of 1:1 [79]; thus, another possibility is that under the circumstances in which the number of phagocyted cells is higher than usual, the apoptotic damage caused to the immune cells could be greater. In the case of ali1Δ and orf19.7590Δ, macrophages had a higher ability to phagocytize and kill them. As these proteins are membrane oxidoreductases, and the mutants are more sensitive to oxidative stress, this result suggests a role for these proteins in avoiding the damage produced by reactive oxidative species inside the macrophages that would be impaired in the mutant cells.
Five out of the eight mutants examined were shown to be of important significance in regulating C. albicans interactions with oral epithelial cells in vitro (ali1Δ, mci4Δ, orf19.287Δ, orf19.3060Δ and orf19.7590Δ). However, while almost all of them showed decreased endocytosis and reduced capacity to damage the oral epithelial cell line, orf19.3060Δ showed decreased endocytosis but a significant increase in cell damage compared with the wild type. As mentioned above, the glycosylation status of the C. albicans cell wall is critical in host–pathogen interaction. Moreover, besides being involved in phagocytosis, it is also critical for the stimulus and regulation of epithelial responses [80]. Moreover, tos1Δ showed high variability in both assays with FaDu cells, showing slightly increased endocytosis and acting similarly to the wild-type strain in the damage assays, suggesting that Tos1 is not essential for adhesion and endocytosis by FaDu cells. As happens with tos1Δ, the orf19.5352Δ mutant showed a high variability in the results. As predicted by their in vitro results in host–cell interactions, no difference was observed in virulence in the mouse model of OPC (Supplemental Fig. S2). Therefore, these proteins de not appear to be necessary for virulence during oropharyngeal candidiasis.
5. Conclusions
The combination of proteomics studies with functional analysis of selected mutant strains allowed the identification of proteins involved in relevant cellular processes such as cell wall maintenance, osmotic and oxidative stress resistance and host–pathogen interplay. More studies are needed to understand the complexity of the C. albicans surface to find new drug targets and biomarkers for Candida infections.
Supplementary Material
Biological significance.
This work describes the identification of surface proteins of C. albicans when grown in the yeast and hypha forms using a cell shaving treatment followed by peptide separation and protein identification using LC-MS/MS methodology. Significant protein differences were observed between the morphologies. The number of detected proteins is the most extensive and complex published up to now using this strategy. This methodology has allowed detecting new proteins related to relevant functions in the host such as cell wall integrity, stress resistance and host-pathogen interaction, verified by phenotypic and virulence assays in vitro and in vivo using 17 mutant strains.
Acknowledgments
This work was supported by BIO2012-31767 from the Ministerio de Economía y Competitividad, PROMPT (S2010/BMD-2414) from the Comunidad Autónoma de Madrid, REIPI, Spanish Network for the Research in Infectious Diseases (RD12/0015/0004), and PRB2-ISCIII (PT13/0001/0038) funded by ISCIII and FEDER. It was also supported by grant R01DE017088 from the National Institutes of Health, USA. A. Gil-Bona was the recipient of a fellowship from Ministerio de Ciencia e Innovación. CM. Parra was the recipient of a fellowship from the program Continuous training for teachers of the Pontificia Javeriana University (Colombia) in conjunction with COLCIENCIAS-LASPAU Harvard University. The proteomic analyses were carried out in the Proteomics Unit UCM-Parque Cientifíco deMadrid, a member of ProteoRed network. Strains and plasmids were obtained from the Fungal Genetics Stock Center (Kansas City, Missouri, USA). The authors would like to thank J. Aguilera and I. García-Barbazán for their helpful technical support in phenotypic assays and SG. Filler lab personnel for their support and help in the works performed in their laboratory. These results are lined up with the Spanish Initiative on the Human Proteome Project (B/DHPP).
References
- 1.Eyerich K, Eyerich S, Hiller J, Behrendt H, Traidl-Hoffmann C. Chronic mucocutaneous candidiasis, from bench to bedside. Eur. J. Dermatol. 2010;20:260–265. doi: 10.1684/ejd.2010.0910. [DOI] [PubMed] [Google Scholar]
- 2.Martins N, Ferreira IC, Barros L, Silva S, Henriques M. Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment. Mycopathologia. 2014;177:223–240. doi: 10.1007/s11046-014-9749-1. [DOI] [PubMed] [Google Scholar]
- 3.Nieto MC, Telleria O, Cisterna R. Sentinel surveillance of invasive candidiasis in Spain: epidemiology and antifungal susceptibility. Diagn. Microbiol. Infect. Dis. 2015;81:34–40. doi: 10.1016/j.diagmicrobio.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Richard ML, Plaine A. Comprehensive analysis of glycosylphosphatidylinositol-anchored proteins in Candida albicans. Eukaryot. Cell. 2007;6:119–133. doi: 10.1128/EC.00297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klis FM, Sosinska GJ, de Groot PW, Brul S. Covalently linked cell wall proteins of Candida albicans and their role in fitness and virulence. FEMS Yeast Res. 2009;9:1013–1028. doi: 10.1111/j.1567-1364.2009.00541.x. [DOI] [PubMed] [Google Scholar]
- 6.Free SJ. Fungal cell wall organization and biosynthesis. Adv. Genet. 2013;81:33–82. doi: 10.1016/B978-0-12-407677-8.00002-6. [DOI] [PubMed] [Google Scholar]
- 7.Nombela C, Gil C, Chaffin WL. Non-conventional protein secretion in yeast. Trends Microbiol. 2006;14:15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Chaffin WL. Candida albicans cell wall proteins. Microbiol. Mol. Biol. Rev. 2008;72:495–544. doi: 10.1128/MMBR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martínez-Lopez R, Monteoliva L, Diez-Orejas R, Nombela C, Gil C. The GPI-anchored protein CaEcm33p is required for cell wall integrity, morphogenesis and virulence in Candida albicans. Microbiology. 2004;150:3341–3354. doi: 10.1099/mic.0.27320-0. [DOI] [PubMed] [Google Scholar]
- 10.Martinez-Lopez R, Park H, Myers CL, Gil C, Filler SG. Candida albicans Ecm33p is important for normal cell wall architecture and interactions with host cells. Eukaryot. Cell. 2006;5:140–147. doi: 10.1128/EC.5.1.140-147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. Yeast wall protein 1 of Candida albicans. Microbiology. 2005;151:1631–1644. doi: 10.1099/mic.0.27663-0. [DOI] [PubMed] [Google Scholar]
- 12.de Groot PW, de Boer AD, Cunningham J, Dekker HL, de Jong L, Hellingwerf KJ, et al. Proteomic analysis of Candida albicans cell walls reveals covalently bound carbohydrate-active enzymes and adhesins. Eukaryot. Cell. 2004;3:955–965. doi: 10.1128/EC.3.4.955-965.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez AI, Castillo L, Garcera A, Elorza MV, Valentin E, Sentandreu R. Role of Pir1 in the construction of the Candida albicans cell wall. Microbiology. 2004;150:3151–3161. doi: 10.1099/mic.0.27220-0. [DOI] [PubMed] [Google Scholar]
- 14.Lan CY, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, et al. Regulatory networks affected by iron availability in Candida albicans. Mol. Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 15.Copping VM, Barelle CJ, Hube B, Gow NA, Brown AJ, Odds FC. Exposure of Candida albicans to antifungal agents affects expression of SAP2 and SAP9 secreted proteinase genes. J. Antimicrob. Chemother. 2005;55:645–654. doi: 10.1093/jac/dki088. [DOI] [PubMed] [Google Scholar]
- 16.Sarthy AV, McGonigal T, Coen M, Frost DJ, Meulbroek JA, Goldman RC. Phenotype in Candida albicans of a disruption of the BGL2 gene encoding a 1,3-beta-glucosyltransferase. Microbiology. 1997;143:367–376. doi: 10.1099/00221287-143-2-367. [DOI] [PubMed] [Google Scholar]
- 17.Pitarch A, Jimenez A, Nombela C, Gil C. Decoding serological response to Candida cell wall immunome into novel diagnostic, prognostic, and therapeutic candidates for systemic candidiasis by proteomic and bioinformatic analyses. Mol. Cell. Proteomics MCP. 2006;5:79–96. doi: 10.1074/mcp.M500243-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Gil-Bona A, Llama-Palacios A, Parra CM, Vivanco F, Nombela C, Monteoliva L, et al. Proteomics unravels extracellular vesicles as carriers of classical cytoplasmic proteins in Candida albicans. J. Proteome Res. 2015;14:142–153. doi: 10.1021/pr5007944. [DOI] [PubMed] [Google Scholar]
- 19.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol. Mol. Biol. Rev. MMBR. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- 21.Lermann U, Morschhauser J. Secreted aspartic proteases are not required for invasion of reconstituted human epithelia by Candida albicans. Microbiology. 2008;154:3281–3295. doi: 10.1099/mic.0.2008/022525-0. [DOI] [PubMed] [Google Scholar]
- 22.Correia A, Lermann U, Teixeira L, Cerca F, Botelho S, da Costa RM, et al. Limited role of secreted aspartyl proteinases Sap1 to Sap6 in Candida albicans virulence and host immune response in murine hematogenously disseminated candidiasis. Infect. Immun. 2010;78:4839–4849. doi: 10.1128/IAI.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer FL, Wilson D, Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Angiolella L, Facchin M, Stringaro A, Maras B, Simonetti N, Cassone A. Identification of a glucan-associated enolase as a main cell wall protein of Candida albicans and an indirect target of lipopeptide antimycotics. J. Infect. Dis. 1996;173:684–690. doi: 10.1093/infdis/173.3.684. [DOI] [PubMed] [Google Scholar]
- 25.Gil-Navarro I, Gil ML, Casanova M, O'Connor JE, Martinez JP, Gozalbo D. The glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is a surface antigen. J. Bacteriol. 1997;179:4992–4999. doi: 10.1128/jb.179.16.4992-4999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urban C, Sohn K, Lottspeich F, Brunner H, Rupp S. Identification of cell surface determinants in Candida albicans reveals Tsa1p, a protein differentially localized in the cell. FEBS Lett. 2003;544:228–235. doi: 10.1016/s0014-5793(03)00455-1. [DOI] [PubMed] [Google Scholar]
- 27.Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 2002;5:366–371. doi: 10.1016/s1369-5274(02)00338-7. [DOI] [PubMed] [Google Scholar]
- 28.Berman J. Morphogenesis and cell cycle progression in Candida albicans. Curr. Opin. Microbiol. 2006;9:595–601. doi: 10.1016/j.mib.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Jacobsen ID, Wilson D, Wachtler B, Brunke S, Naglik JR, Hube B. Candida albicans dimorphism as a therapeutic target. Expert Rev. Anti-Infect. Ther. 2012;10:85–93. doi: 10.1586/eri.11.152. [DOI] [PubMed] [Google Scholar]
- 31.Ebanks RO, Chisholm K, McKinnon S, Whiteway M, Pinto DM. Proteomic analysis of Candida albicans yeast and hyphal cell wall and associated proteins. Proteomics. 2006;6:2147–2156. doi: 10.1002/pmic.200500100. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Gomariz M, Perumal P, Mekala S, Nombela C, Chaffin WL, Gil C. Proteomic analysis of cytoplasmic and surface proteins from yeast cells, hyphae, and biofilms of Candida albicans. Proteomics. 2009;9:2230–2252. doi: 10.1002/pmic.200700594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heilmann CJ, Sorgo AG, Siliakus AR, Dekker HL, Brul S, de Koster CG, et al. Hyphal induction in the human fungal pathogen Candida albicans reveals a characteristic wall protein profile. Microbiology. 2011;157:2297–2307. doi: 10.1099/mic.0.049395-0. [DOI] [PubMed] [Google Scholar]
- 34.Sosinska GJ, de Koning LJ, de Groot PW, Manders EM, Dekker HL, Hellingwerf KJ, et al. Mass spectrometric quantification of the adaptations in the wall proteome of Candida albicans in response to ambient pH. Microbiology. 2011;157:136–146. doi: 10.1099/mic.0.044206-0. [DOI] [PubMed] [Google Scholar]
- 35.Vialas V, Perumal P, Gutierrez D, Ximenez-Embun P, Nombela C, Gil C, et al. Cell surface shaving of Candida albicans biofilms, hyphae, and yeast form cells. Proteomics. 2012;12:2331–2339. doi: 10.1002/pmic.201100588. [DOI] [PubMed] [Google Scholar]
- 36.Hernáez ML, Ximenez-Embún P, Martínez-Gomariz M, Gutierrez- Blazquez MD, Nombela C, Gil C. Identification of Candida albicans exposed surface proteins in vivo by a rapid proteomic approach. J. Proteomics. 2010;73:1404–1409. doi: 10.1016/j.jprot.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 37.Gillum AM, Tsay EYH, Kirsch DR. Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxilase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. 1984;198:179–182. doi: 10.1007/BF00328721. [DOI] [PubMed] [Google Scholar]
- 38.Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCluskey K, Wiest A, Plamann M. The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 2010;35:119–126. doi: 10.1007/s12038-010-0014-6. [DOI] [PubMed] [Google Scholar]
- 40.Lee KL, Buckley HR, Campbell CC. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia. 1975;13:148–153. doi: 10.1080/00362177585190271. [DOI] [PubMed] [Google Scholar]
- 41.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 42.Zybailov BL, Florens L, Washburn MP. Quantitative shotgun proteomics using a protease with broad specificity and normalized spectral abundance factors. Mol. BioSyst. 2007;3:354–360. doi: 10.1039/b701483j. [DOI] [PubMed] [Google Scholar]
- 43.Fernández-Arenas E, Cabezón V, Bermejo C, Arroyo J, Nombela C, Diez-Orejas R, et al. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol. Cell. Proteomics. 2007;6:460–478. doi: 10.1074/mcp.M600210-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AA, EE J, et al. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell. Microbiol. 2005;7:499–510. doi: 10.1111/j.1462-5822.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- 45.Phan QT, Belanger PH, Filler SG. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 2000;68:3485–3490. doi: 10.1128/iai.68.6.3485-3490.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solis NV, Filler SG. Mouse model of oropharyngeal candidiasis. Nat. Protoc. 2012;7:637–642. doi: 10.1038/nprot.2012.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vialas V, Sun Z, Loureiro y Penha CV, Carrascal M, Abian J, Monteoliva L, et al. A Candida albicans PeptideAtlas. J. Proteomics. 2014;97:62–68. doi: 10.1016/j.jprot.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pardo M, Monteoliva L, Vazquez P, Martinez R, Molero G, Nombela C, et al. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology. 2004;150:4157–4170. doi: 10.1099/mic.0.26924-0. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz C, Cid VJ, Lussier M, Molina M, Nombela C. A large-scale sonication assay for cell wall mutant analysis in yeast. Yeast. 1999;15:1001–1008. doi: 10.1002/(SICI)1097-0061(199907)15:10B<1001::AID-YEA400>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 50.Kamai Y, Kubota M, Kamai Y, Hosokawa T, Fukuoka T, Filler SG. New model of oropharyngeal candidiasis in mice. Antimicrob. Agents Chemother. 2001;45:3195–3197. doi: 10.1128/AAC.45.11.3195-3197.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pardo M, Monteoliva L, Pla J, Sanchez M, Gil C, Nombela C. Two-dimensional analysis of proteins secreted by Saccharomyces cerevisiae regenerating protoplasts: a novel approach to study the cell wall. Yeast. 1999;15:459–472. doi: 10.1002/(SICI)1097-0061(199904)15:6<459::AID-YEA387>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 52.Pitarch A, Sanchez M, Nombela C, Gil C. Sequential fractionation and two dimensional gel analysis unravels the complexity of the dimorphic fungus Candida albicans cell wall proteome. Mol. Cell. Proteomics. 2002;1:967–982. doi: 10.1074/mcp.m200062-mcp200. [DOI] [PubMed] [Google Scholar]
- 53.Castillo L, Calvo E, Martinez AI, Ruiz-Herrera J, Valentin E, Lopez JA, et al. A study of the Candida albicans cell wall proteome. Proteomics. 2008;8:3871–3881. doi: 10.1002/pmic.200800110. [DOI] [PubMed] [Google Scholar]
- 54.Insenser MR, Hernaez ML, Nombela C, Molina M, Molero G, Gil C. Gel and gel-free proteomics to identify Saccharomyces cerevisiae cell surface proteins. J. Proteomics. 2010;73:1183–1195. doi: 10.1016/j.jprot.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Olaya-Abril A, Jimenez-Munguia I, Gomez-Gascon L, Rodriguez-Ortega MJ. Surfomics: shaving live organisms for a fast proteomic identification of surface proteins. J. Proteomics;2014;97:164–176. doi: 10.1016/j.jprot.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 56.Bailey DA, Feldmann PJF, Bovey M, Gow NAR, Brown AJP. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoyer LL, Payne TL, Hecht JE. Identification of Candida albicans ALS2 and ALS4 and localization of als proteins to the fungal cell surface. J. Bacteriol. 1998;180:5334–5343. doi: 10.1128/jb.180.20.5334-5343.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martchenko M, Alarco AM, Harcus D, Whiteway M. Superoxide dismutases in Candida albicans: transcriptional regulation and functional characterization of the hyphal-induced SOD5 gene. Mol. Biol. Cell. 2004;15:456–467. doi: 10.1091/mbc.E03-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan QT, et al. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2006;2:e63. doi: 10.1371/journal.ppat.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Argimon S, Wishart JA, Leng R, Macaskill S, Mavor A, Alexandris T, et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell. 2007;6:682–692. doi: 10.1128/EC.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr. Opin. Microbiol. 2009;12:365–370. doi: 10.1016/j.mib.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Muhlschlegel FA, Fonzi WA. PHR2 of Candida albicans encodes a functional homolog of the pH-regulated gene PHR1 with an inverted pattern of pH-dependent expression. Mol. Cell. Biol. 1997;17:5960–5967. doi: 10.1128/mcb.17.10.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fonzi WA. PHR1 and PHR2 of Candida albicans encode putative glycosidases required for proper cross-linking of beta-1,3- and beta-1,6-glucans. J. Bacteriol. 1999;181:7070–7079. doi: 10.1128/jb.181.22.7070-7079.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sorgo AG, Heilmann CJ, Dekker HL, Bekker M, Brul S, de Koster CG, et al. Effects of fluconazole on the secretome, the wall proteome, and wall integrity of the clinical fungus Candida albicans. Eukaryot. Cell. 2011;10:1071–1081. doi: 10.1128/EC.05011-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heilmann CJ, Sorgo AG, Mohammadi S, Sosinska GJ, de Koster CG, Brul S, et al. Surface stress induces a conserved cell wall stress response in the pathogenic fungus Candida albicans. Eukaryot. Cell. 2013;12:254–264. doi: 10.1128/EC.00278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol. Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 67.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, et al. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell. Microbiol. 2008;10:2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niimi M, Tokunaga M, Nakayama H. Regulation of mannitol catabolism in Candida albicans: evidence for cyclic AMP-independent glucose effect. J. Med. Vet. Mycol. 1986;24:211–217. doi: 10.1080/02681218680000311. [DOI] [PubMed] [Google Scholar]
- 69.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5:e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monteoliva L, Martinez-Lopez R, Pitarch A, Hernaez ML, Serna A, Nombela C, et al. Quantitative proteome and acidic subproteome profiling of Candida albicans yeast-to-hypha transition. J. Proteome Res. 2011;10:502–517. doi: 10.1021/pr100710g. [DOI] [PubMed] [Google Scholar]
- 71.Pardo M, Ward M, Bains S, Molina M, Blackstock W, Gil C, et al. A proteomic approach for the study of Saccharomyces cerevisiae cell wall biogenesis. Electrophoresis. 2000;21:3396–3410. doi: 10.1002/1522-2683(20001001)21:16<3396::AID-ELPS3396>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 72.Nobile CJ, Bruno VM, Richard ML, Davis DA, Mitchell AP. Genetic control of chlamydospore formation in Candida albicans. Microbiology. 2003;149:3629–3637. doi: 10.1099/mic.0.26640-0. [DOI] [PubMed] [Google Scholar]
- 73.McKenzie CG, Koser U, Lewis LE, Bain JM, Mora-Montes HM, Barker RN, et al. Contribution of Candida albicans cell wall components to recognition by and escape from murine macrophages. Infect. Immun. 2010;78:1650–1658. doi: 10.1128/IAI.00001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sorgo AG, Heilmann CJ, Dekker HL, Brul S, de Koster CG, Klis FM. Mass spectrometric analysis of the secretome of Candida albicans. Yeast. 2010;27:661–672. doi: 10.1002/yea.1775. [DOI] [PubMed] [Google Scholar]
- 75.Sorgo AG, Heilmann CJ, Brul S, de Koster CG, Klis FM. Beyond the wall: Candida albicans secret(e)s to survive. FEMS Microbiol. Lett. 2013;338:10–17. doi: 10.1111/1574-6968.12049. [DOI] [PubMed] [Google Scholar]
- 76.Vargas G, Rocha JD, Oliveira DL, Albuquerque PC, Frases S, Santos SS, et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell. Microbiol. 2015;17:389–407. doi: 10.1111/cmi.12374. [DOI] [PubMed] [Google Scholar]
- 77.Martin R, Albrecht-Eckardt D, Brunke S, Hube B, Hunniger K, Kurzai O. A core filamentation response network in Candida albicans is restricted to eight genes. PLoS One. 2013;8:e58613. doi: 10.1371/journal.pone.0058613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reales-Calderon JA, Aguilera-Montilla N, Corbi AL, Molero G, Gil C. Proteomic characterization of human pro-inflammatory M1 and anti-inflammatory M2 macrophages and their response to Candida albicans. Proteomics. 2014;14 doi: 10.1002/pmic.201300508. [DOI] [PubMed] [Google Scholar]
- 79.Reales-Calderon JA, Sylvester M, Strijbis K, Jensen ON, Nombela C, Molero G, et al. Candida albicans induces pro-inflammatory and anti-apoptotic signals in macrophages as revealed by quantitative proteomics and phosphoproteomics. J. Proteomics. 2013;9:106–135. doi: 10.1016/j.jprot.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 80.Wagener J, Weindl G, de Groot PW, de Boer AD, Kaesler S, Thavaraj S, et al. Glycosylation of Candida albicans cell wall proteins is critical for induction of innate immune responses and apoptosis of epithelial cells. PLoS One. 2012;7:e50518. doi: 10.1371/journal.pone.0050518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.