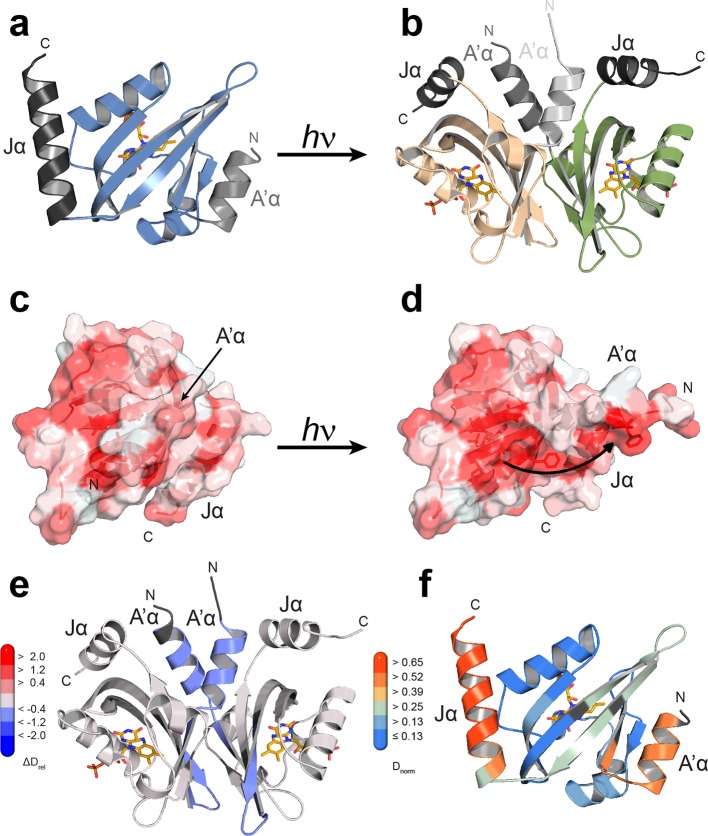
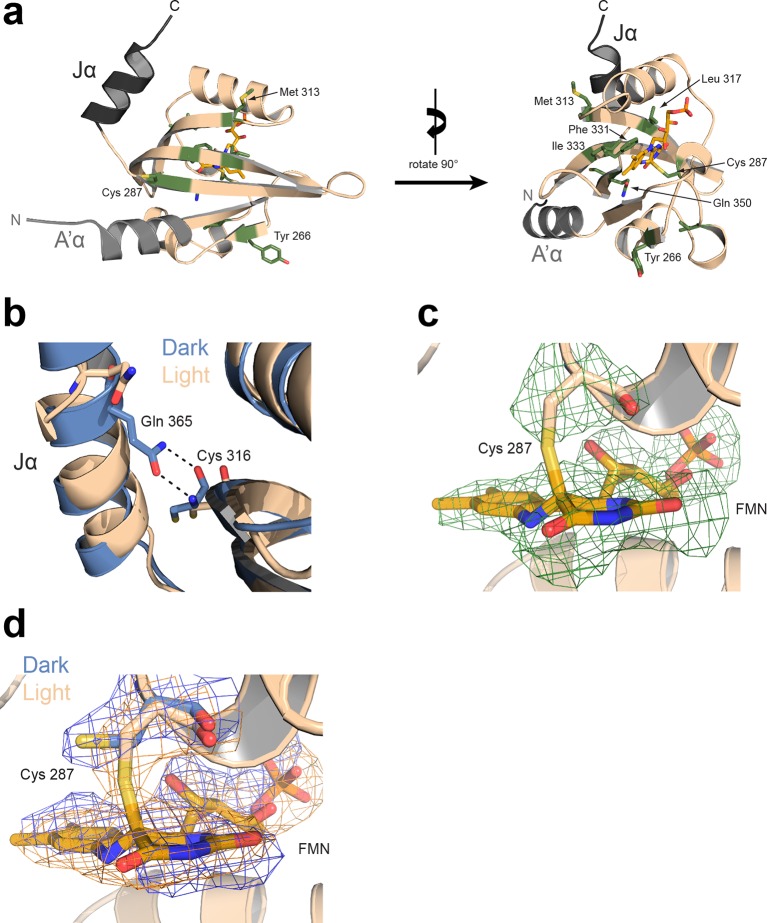
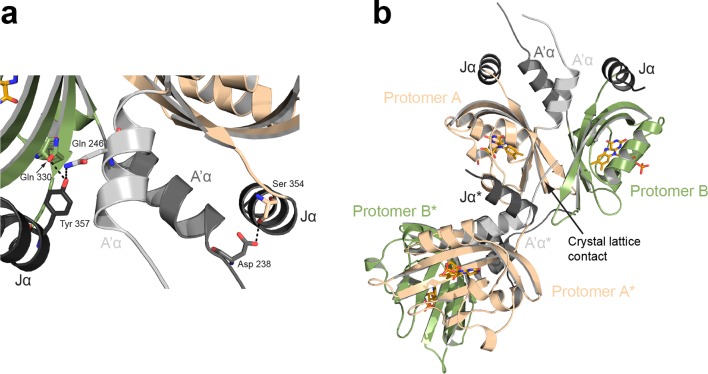
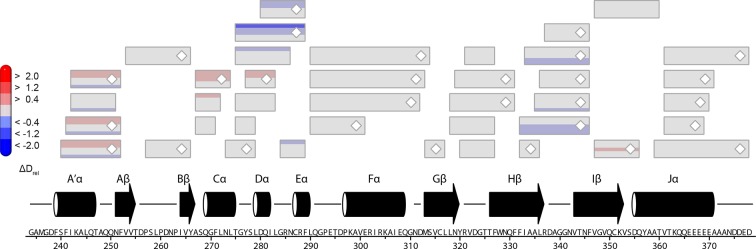
Figure 4. Structural characterization of PtAu1aLOV in its dark and light state.
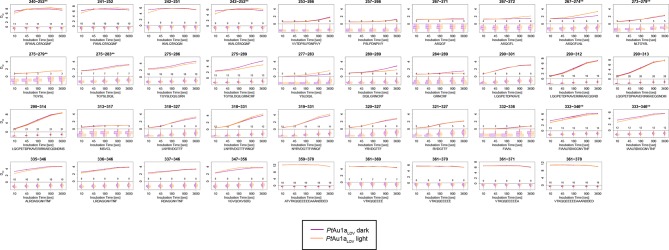
(a) Crystal structure of the PtAu1aLOV dark state monomer with the N- and C-terminal A'α and Jα helices flanking the LOV core colored in light gray and dark gray, respectively. (b) Blue light illumination induces formation of a parallel PtAu1aLOV dimer. (c) In the dark, A'α covers the hydrophobic dimerization site on the LOV β-sheet. (d) Illumination results in a release of A'α from the LOV β-sheet and exposes the dimerization site. The PtAu1aLOV molecules in (c and d) are colored according to the Eisenberg hydrophobicity scale (Eisenberg, et al., 1984). Reddish regions correspond to high and white regions to low hydrophobicity. (e) The PtAu1aLOV light state dimer colored according to differences in deuterium incorporation in the dark and light state after 10 s of labeling. Shades of red and blue correspond to regions with increased and decreased deuterium uptake in the light, respectively. A peptide map that shows the differences in relative deuteration of dark and light experiments for all time points is shown in (Figure 4—figure supplement 3). All evaluated peptides for PtAu1aLOV and their individual deuteration plots are shown in (Figure 4—figure supplement 4). (f) PtAu1aLOV dark state monomer colored according to deuterium incorporation in the dark after 10 s labeling. Elements in (e) and (f) colored in dark gray represent regions that are not covered by peptides generated by pepsin digestion. Since rapid back-exchange of the two N-terminal residues prevents precise measurement of deuterium incorporation, these residues of all peptides are shown in dark gray, if not covered by an overlapping peptide. LOV, light-oxygen-voltage.