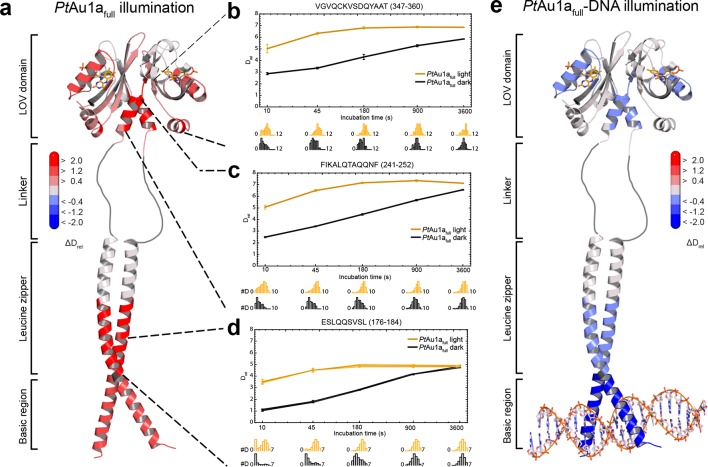
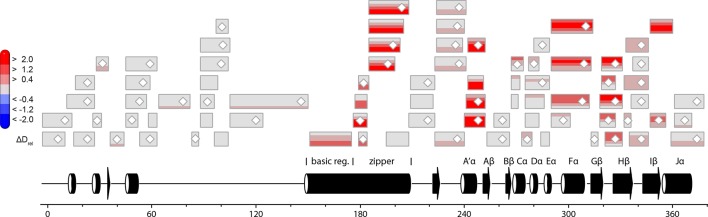
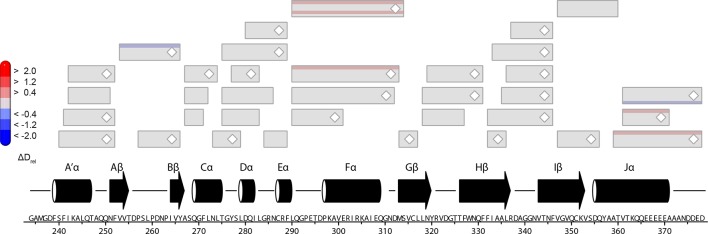
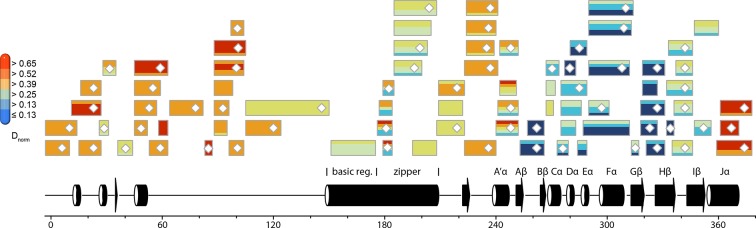
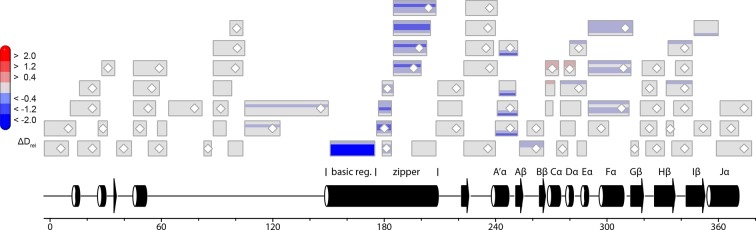
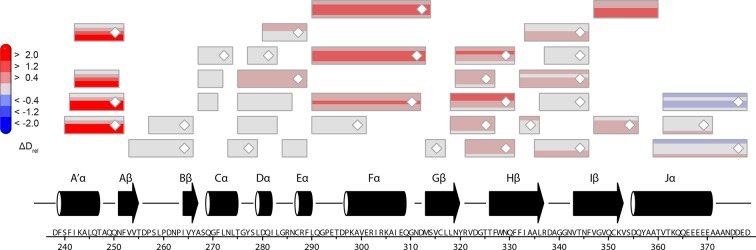
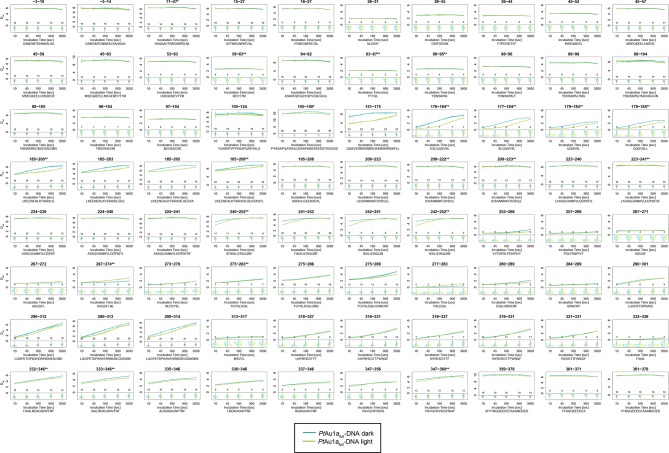
Figure 5. HDX-MS data of PtAu1afull in the absence and presence of DNA.
(a) Changes in deuterium incorporation of PtAu1afull mapped onto the structure of the PtAu1aLOV light state dimer and a model of the bZIP domain. (b–d) Deuterium uptake plots of Jα-Iβ, A'α and leucine zipper peptides with Drel plotted against the labeling time for three independent experiments. The estimated abundance distribution of individual deuterated species is shown at the bottom. (e) Differences in deuterium incorporation of PtAu1afull in the dark and light state in the presence of DNA mapped onto the PtAu1aLOV light state dimer and a model of the bZIP domain. All evaluated peptides for PtAu1afull in the absence and presence of DNA and their individual deuteration plots are shown in Figure 5—figure supplement 6 and Figure 5—figure supplement 7, respectively. HDX-MS, hydrogen/deuterium-exchange coupled to mass spectrometry; LOV, light-oxygen-voltage.