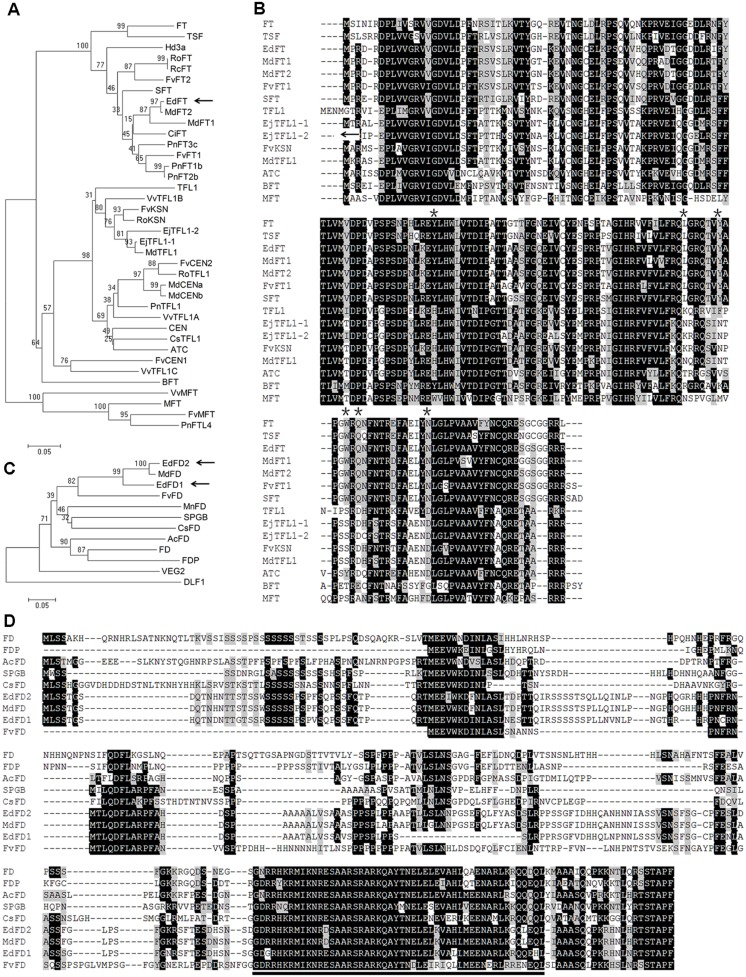
FIGURE 1.
Sequence analysis of EdFT and EdFDs. (A) Phylogenetic analysis of plant PEBP family proteins. The accession number of each gene is as follows: FT (Arabidopsis thaliana, AT1G65480), TFL1 (AT5G03840), TSF (AT4G20370), MFT (AT1G18100), BFT (AT5G62040), ATC (AT2G27550), SFT (Solanum lycopersicum, AY186735), EjTFL1-1 (Eriobotrya japonica, AB162045), EjTFL1-2 (AB162051), MdFT1 (Malus× domestica, AB161112), MdFT2 (AB458504), MdTFL1 (AB052994), MdCENa (AB366641), MdCENb (AB366642), FvFT1 (Fragariavesca, JN172098), FvFT2 (XM_004297225), FvCEN1 (XM_004308059), FvCEN2 (XM_004291562), FvKSN (HQ378595), FvMFT (XM_004299493), RoFT (Rosa luciae, FM999826), RoTFL1 (FM999796), RoKSN (HQ174211), RcFT (Rosa chinensis, FR729041), CiFT (Citrus unshiu, AB027456), PnFT1b (Populus nigra, AB161109), PnFT2b (AB161108), PnFT3c (AB161107), PnTFL1 (AB369067), PnFTL4 (AB181241), Hd3a (Oryza sativa, AB052941), CEN (Antirrhinum, S81193), CsTFL1 (Chrysanthemum seticuspe, AB839767), VvTFL1A (Vitis vinifera, DQ871591), VvTFL1B (DQ871592), VvTFL1C (DQ871593), VvMFT (DQ871594). Arrows indicate the EdFT gene. (B) Amino acid sequence alignment of several plant PEBP family proteins showed in (A). Amino acid residues in black and gray represent 100 and 50% similarity respectively. The asterisks indicate the critical amino acid positions. (C) Phylogenetic analysis of plant FD proteins. The accession number of each gene is as follows: FD (AT4G35900), FDP (AT2G17770), AcFD (Actinidia chinensis, JX417425), SPGB (Solanum lycopersicum, EF136919), MnFD (Morus notabilis, XM_010115002), VEG2 (Pisum sativum, KP739949), CsFD (Chrysanthemum seticuspe, AB839769), DLF1 (Zea mays, NM_001112492), MdFD (Malus × domestica, XM_008339760), FvFD (Fragaria vesca, XM_004289026). Arrows indicate the EdFD1/2 genes. (D) Amino acid sequence alignment of several plant FD proteins showed in (C). Amino acid residues in black and gray represent 100 and 50% similarity, respectively. The bZIP domain is underlined.