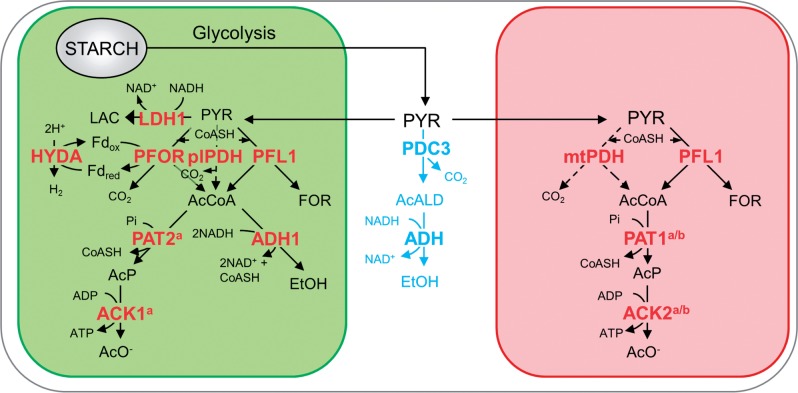
Fig. 9.
Proposed model of the subcellular localization of fermentative pathways in C. reinhardtii. Compartments within the cell are displayed: chloroplast (green), mitochondrion (red), cytoplasm (white). Highlighted are aerobic pathways (dashed line) and the putative cytoplasmic pyruvate degradation pathway (light blue). Abbreviations: Fdox, ferredoxin (oxidized); Fdred, ferredoxin (reduced); AcALD, acetaldehyde; AcCoA, acetyl-CoA; AcP, acetyl-phosphate; CoASH, acetyl-CoA; EtOH, ethanol; FOR, formate; AcO–, acetate; LAC, lactate; PYR, pyruvate. The annotated genes are: ACK1, acetate kinase 1 (Cre09.g396700); ACK2, acetate kinase 2 (Cre17.g709850); ADH1, acetaldehyde/alcohol dehydrogenase; HYDA1, [Fe–Fe]-hydrogenase (Cre20.g758200); LDH1, d-lactate dehydrogenase (Cre07.g324550); PAT2, phosphate acetyltransferase 2 (Cre09.g396650); plPDH, plastid pyruvate dehydrogenase complex; mtPDH, mitochondrial pyruvate dehydrogenase complex; PDC3, pyruvate decarboxylase 3 (Cre03.g165700); PFL1, pyruvate formate lyase (Cre01.g044800); PFOR, pyruvate:ferredoxin oxidoreductase (Cre11.g473950). Where specific designations have not been given, there are multiple isoforms present in C. reinhardtii and the precise enzyme involved has not yet been verified. aAs localized by Terashima et al. (2010). bAs identified by Atteia et al. (2009). A number of additional pathways are thought to be active in production of intracellular metabolites but were not analyzed here and have been excluded for simplicity; see Subramanian et al. (2014) for details. Additionally the glycolytic pathway has been omitted; see Klein (1986) for details of partitioning between the chloroplast and cytoplasm.