FIGURE 1.
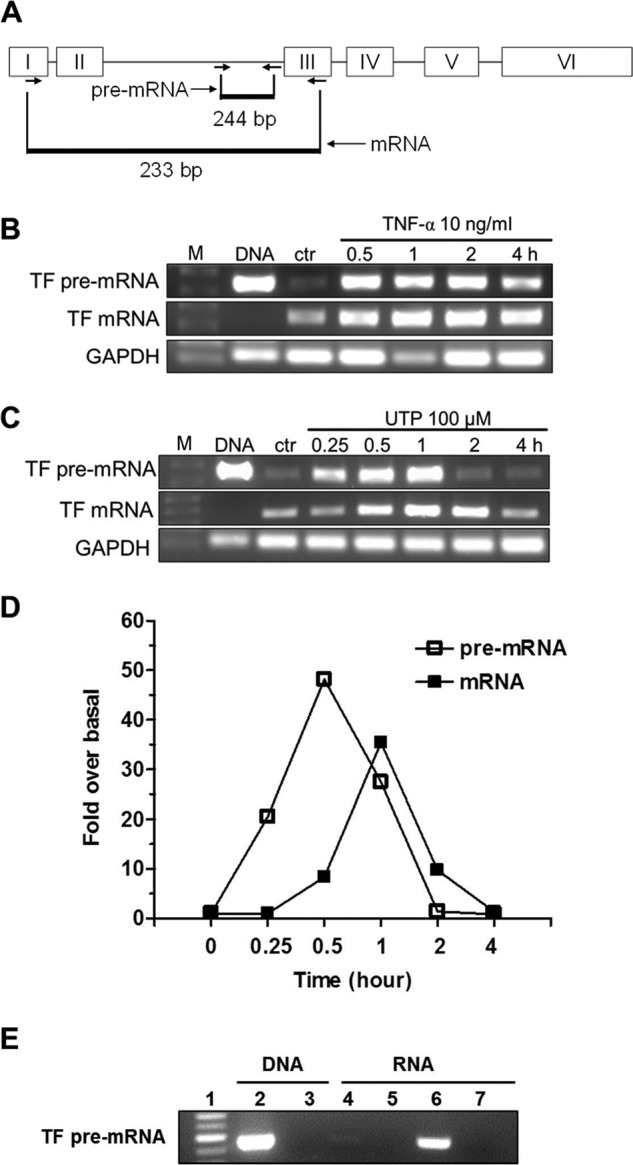
P2Y2R activation promotes TF gene transcription in HCAEC. Two pairs of primers were designed to quantify human TF mature mRNA and pre-mRNA (hnRNA) by RT-PCR, respectively. The relative position of primers to the TF gene is depicted in A. Representative RT-PCR gel images show time-dependent effect of TNF-α (B) or UTP (C) on the expression of TF pre-mRNA and mature mRNA. In both panels, the 1st lanes are DNA markers (M); 2nd lanes are PCR products of DNA as a positive control; 3rd lanes are from total RNA samples made from HCAEC without stimulation. The rest of the lanes reflect time course changes caused by TNF-α (B) and UTP (C), respectively. GAPDH was used as a control. Quantitative data on time course change of TF mRNA and pre-mRNA in response to UTP (100 μm) treatment were determined by real time RT-PCR (D). For clarity, error bars and markers for statistical analysis were omitted (n = 3). E, evidence of complete clearance of genomic DNA by DNase treatment in DNA or total RNA samples. Lane 1, standard DNA ladder 100 bp; lane 2, PCR product amplified from 200 ng of genomic DNA prepared from HCAEC; lane 3, PCR product amplified from 200 ng of genomic DNA treated with DNase; lane 4, product from RT-PCR of total RNA prepared from HCAEC without UTP stimulation; lane 5, PCR product amplified from non-reverse-transcribed total RNA prepared from HCAEC without UTP stimulation; lane 6, product from RT-PCR of total RNA prepared from HCAEC treated with 100 μm UTP for 1 h; lane 7, PCR product amplified from non-reverse-transcribed total RNA prepared from HCAEC treated with 100 μm UTP for 1 h (n = 3).
