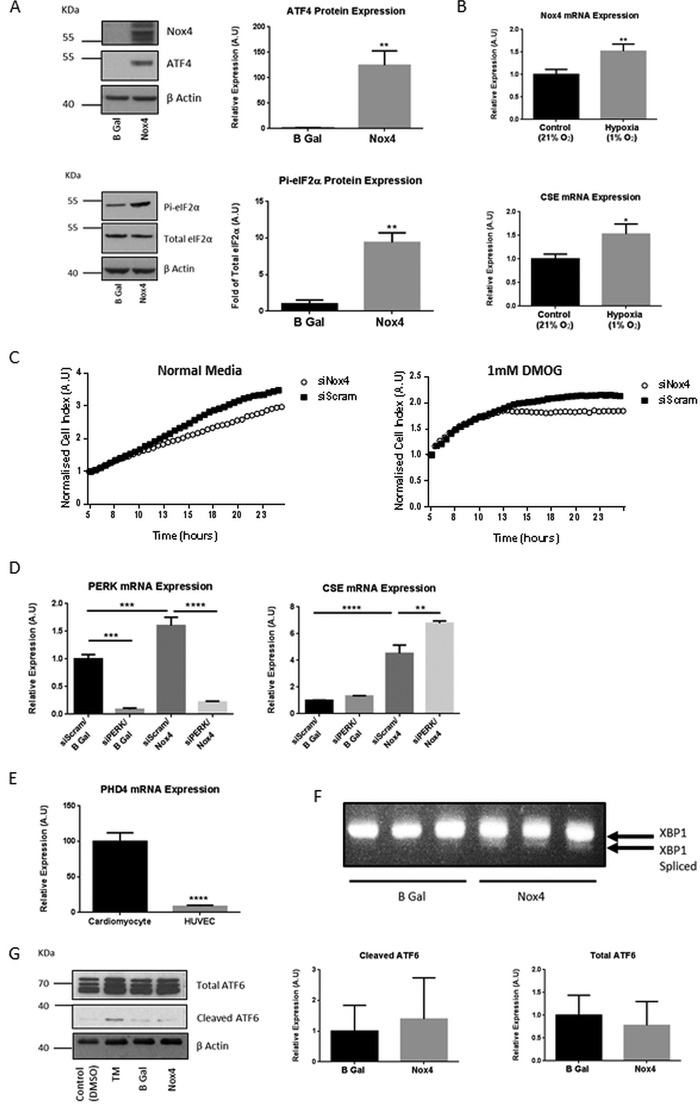
FIGURE 4.
Nox4 elicited a cellular stress response. A, representative Western blots and quantitative densitometric analysis of ATF4, total eIF2α and phosphorylated eIF2α protein (Pi-eIF2α) in HUVECs after 24-h Nox4 or β-gal (B Gal) overexpression. A.U., absorbance units. B, QPCR analyses of Nox4 and CSE mRNA expression in HUVEC after 1 h of incubation under atmospheric (Control (21%)) or hypoxic (Hypoxia (1% O2) conditions. C, HUVEC proliferation assessed after treatment with siRNA targeted to Nox4 (siNox4) or control siRNA (siScram) for 24 h. Cells were then seeded at equal densities onto E-plates (ACEA), and respective cell index was subsequently measured on an xCELLigence real time cell analyzer for 24 h. Cells were cultured in normal media or in media supplemented with 1 mm dimethyloxalylglycine (DMOG), administered 5 h after plating. The cell indices were normalized at the time point of compound administration in all cases (5 h). D, QPCR analyses of PERK and CSE mRNA expression in HUVECs after 48 h of treatment with siRNA targeted to PERK (siPERK) or control siRNA (siScram) together with 24 h Nox4 or β-gal overexpression as indicated. E, QPCR analyses demonstrating relative PHD4 mRNA expression in (rat) neonatal cardiomyocytes and HUVEC. F, PCR analysis of spliced XBP1 mRNA in HUVEC splicing after 24-h Nox4 or β-gal overexpression as indicated. G, representative Western blot and corresponding densitometric analysis for cleaved and total ATF6 protein expression in HUVECs after 24-h Nox4 or β-gal overexpression as indicated. HUVECs treated with tunicamycin (TM; 2 μg/ml) for 2 h compared with control (vehicle; DMSO) served as a positive control. All data were normalized to β-actin mRNA or protein expression apart from protein that was normalized to total eIF2α protein. n = 3 in all cases, *, p < 0.05; **, p < 0.01; ***; p < 0.001; ****, p < 0.0001.