Abstract
Carney triad (CT) is a rare condition with synchronous or metachronous occurrence of gastrointestinal stromal tumors (GISTs), paragangliomas (PGLs), and pulmonary chondromas in a patient. In contrast to Carney–Stratakis syndrome (CSS) and familial PGL syndromes, no germline or somatic mutations in the succinate dehydrogenase (SDH) complex subunits A, B, C, or D have been found in most tumors and/or patients with CT. Nonetheless, the tumors arising among patients with CT, CSS, or familial PGL share a similar morphology with loss of the SDHB subunit on the protein level. For the current study, we employed massive parallel bisulfite sequencing to evaluate DNA methylation patterns in CpG islands in proximity to the gene loci of all four SDH subunits. For the first time, we report on a recurrent aberrant dense DNA methylation at the gene locus of SDHC in tumors of patients with CT, which was not present in tumors of patients with CSS or PGL, or in sporadic GISTs with KIT mutations. This DNA methylation pattern was correlated to a reduced mRNA expression of SDHC, and concurrent loss of the SDHC subunit on the protein level. Collectively, these data suggest epigenetic inactivation of the SDHC gene locus with functional impairment of the SDH complex as a plausible alternate mechanism of tumorigenesis in CT.
Keywords: Carney triad, SDH complex, SDHC, GIST, paraganglioma
Introduction
Carney triad (CT) is a rare condition with synchronous or metachronous occurrence in a patient of three tumor entities, namely gastric gastrointestinal stromal tumor (GIST), paraganglioma (PGL), and pulmonary chondroma (Carney et al. 1977). The disease has a striking predilection for young females, for reasons that remain unknown. Two of the tumor components, GIST and PGL, are shared by the Carney–Stratakis syndrome (CSS), which affects both genders during childhood and adolescence (Carney & Stratakis 2002). However, in contrast to the nonhereditary CT, CSS follows an autosomal-dominant inheritance pattern (Carney & Stratakis 2002). Inactivating germline mutations within one of the four subunits A, B, C, or D of the succinate dehydrogenase (SDH) complex of the respiratory chain have been identified in CSS patients, with concurrent somatic inactivation of the second allele by mutation or deletion in the tumor (McWhinney et al. 2007, Pasini et al. 2008). Moreover, SDH germline mutations in combination with somatic inactivation of the second allele are the cause of familial PGL syndromes PGL1 (SDHD), PGL3 (SDHC), and PGL4 (SDHB) (Pasini & Stratakis 2009, Welander et al. 2011).
In contrast to CSS and familial PGL, SDH mutations have not been reported in the germline and/or tumors DNA of most patients with CT (Matyakhina et al. 2007). Given the shared loss of SDHB protein in tumors among all three syndromes (van Nederveen et al. 2009), we hypothesized epigenetic silencing of SDH genes as an alternate mechanism in tumors occurring in CT patients. In the current study, we performed broad, high-resolution, and quantitative assessment of DNA methylation in the proximity of transcriptional start sites of all four genes encoding the SDH subunits in tumors from four CT patients who did not have any SDH-coding sequence abnormalities. The DNA methylation patterns were compared with tumors from a patient with CSS, a patient with PGL1 as well as sporadic GISTs harboring mutations in the KIT receptor. Notably, extensive DNA hypermethylation was detected at the gene locus of SDHC in all tumors from the CT patients, while virtually no methylation was detectable in any of the other tumor specimens. mRNA expression of all four SDH subunits A, B, C, and D was determined by quantitative PCR (qPCR), and a significant downregulation of SDHC on mRNA level in the CT tumors was observed, which was in contrast to a virtually equal expression of all four SDH subunits in the other tumor samples. Both SDHB and SDHC subunits were absent at the protein level in the tumors from the CT patients. Collectively, these data demonstrate that DNA methylation of the SDHC gene locus is a recurrent and specific event in tumors of CT patients, which suggests SDHC downregulation by epigenetic inactivation as a plausible tumor-initiating event.
Subjects and methods
Patient cohort
The current study comprised a selection of tumor tissues and non-neoplastic tissues from four patients with CT, one patient with germline SDHB mutation in the context of CSS, one patient with SDHD germline mutation in the context of PGL1, as well as from five sporadic GISTs with somatic activating KIT mutations (Table 1). Among the four CT patients, three presented with the complete triad, while the second patient presented with only two components. The first patient (CT-1) is a 53-year-old female who presented with PGL of the carotid body (Fig. 1). Previously, she had resection of a gastric mesenchymal tumor at the age of 25 years, retrospectively a gastric GIST. Computed topography scans revealed three pulmonary nodules and a gastric GIST. The cervical PGL and the gastric GIST were resected, while the pulmonary nodules consistent with pulmonary chondromas on biopsy were not according to their asymptomatic presentation. The second patient (CT-2) presented at the age of 15 years with gastric GIST with liver metastases and synchronous PGL of the aorticopulmonary ganglion. The third patient (CT-3) is a 46-year-old female. She had GIST tumor resection at the age of 34 years, and presented with a PGL and a pulmonary chondroma in the subsequent years. The fourth patient (CT-4) is a 21-year-old female. She was diagnosed with GIST, PGLs in the abdomen and the carotid body, and one pulmonary chondroma, at the age of 11. The four patients' relatives revealed no components of CT. All tumors associated with CT, CSS, or PGL1 were negative for SDHB by immunohistochemistry, while all sporadic KIT-mutated GISTs displayed the normal SDHB immunostaining with characteristic granular pattern. Mutation analysis of KIT exons 9, 11, 13, and 17; PDGFRA exons 12, 14, and 18 as well as of the complete coding sequence of all four SDH subunits A, B, C, and D was carried out, as described previously (McWhinney et al. 2007, Merkelbach-Bruse et al. 2010, Rattenberry et al. 2013). This study was approved by the Ethics Committee of the Friedrich-Alexander University Erlangen-Nuremberg, Germany (no. 215_12 B, 19.09.2012), and/or by the Institutional Review Boards of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, USA. A written informed consent was obtained from all patients.
Table 1.
Summary of clinicopathological data
| Patient | Sex/age (years)a | Germline mutations | Somatic mutations | SDHB immunostaining in the tumors | GIST location | PGL location | Pulmonary chondroma (n) |
|---|---|---|---|---|---|---|---|
| Patients with CT | |||||||
| CT-1 | F/25 | WTb | WTb | Negative | Stomach (25 years) and stomach (53 years) | Carotid body | Three |
| CT-2 | F/15 | WTb | WTb | Negative | Stomach with liver metastasis | Aortico-pulmonary ganglion | None |
| CT-3 | F/34 | WTb | WTb | Negative | Stomach with liver metastasis | Upper abdomen | One |
| CT-4 | F/11 | WTb | WTb | Negative | Stomach | Multiple in abdomen and carotid body | One |
| Patients with germline SDH mutations | |||||||
| CSS | F/70 | SDHB c.220G>A; p.Asp74Asn | Loss of SDHB WT allele | Negative | Jejunum | None | None |
| PGL1 | F/36 | SDHD c.209G>T; p.Arg70Met | Loss of SDHD WT allele | Negative | None | Carotid body | None |
| Patients with sporadic KIT-mutated GISTs | |||||||
| KIT-1 | F/80 | Not tested | KIT c.1676T>A; p.Val559Asp | Positive | Stomach | None | None |
| KIT-2 | M/63 | Not tested | KIT c.1672_1692del; p.Lys558_Asn564del | Positive | Stomach | None | None |
| KIT-3 | F/62 | Not tested | KIT c.1669_1674del; p.Trp557_Lys558del | Positive | Duodenum | None | None |
| KIT-4 | M/83 | Not tested | KIT c.1676T>A; p.Val559Asp | Positive | Jejunum | None | None |
| KIT-5 | M/75 | Not tested | KIT c.1679T>A; p.Val560Asp | Positive | Stomach | None | None |
Age at first presentation.
No mutation in the coding regions of SDHA, SDHB, SDHC, and SDHD; no mutations in KIT exons 9, 11, 13, and 17; PDGFRA exons 12, 14, and 18. CT, Carney triad.
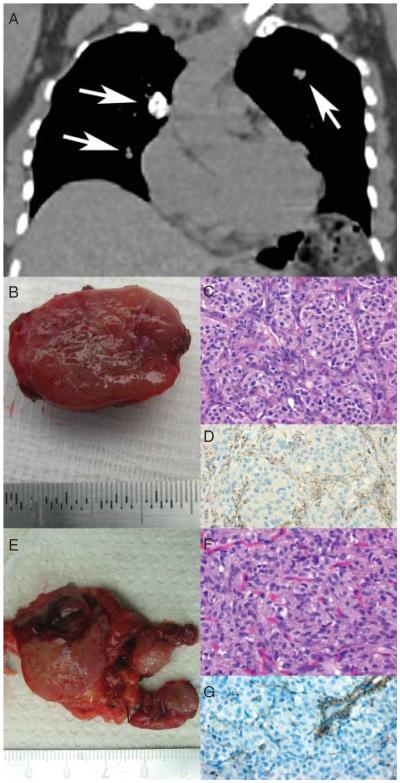
Figure 1.
Clinicopathological presentation of a patient with complete form of Carney triad (CT-1). (A) Coronal reconstruction of thoracic CT scan displaying three pulmonary nodules (arrows) consistent with pulmonary chondroma on biopsy. (B) Gross specimen of the resected PGL of the carotid body. (C) Histologic examination reveals the typical `Zellballen' morphology of PGL (H&E, ×400). (D) SDHB immunostaining is absent in the tumor cells but preserved in the surrounding sustentacular cells and endothelial cells (SDHB, ×400). (E) Gross specimen of the gastric GIST. Note the multinodular appearance. (F) Histologic examination displays a characteristic epithelioid morphology (H&E, ×400). (G) SDHB immunostaining is lost in the tumor cells but preserved in the endothelial cells.
DNA isolation and bisulfite conversion
DNA was extracted from the samples using the DNeasy Blood and Tissue Kit (Qiagen), as suggested by the manufacturer. DNA concentration was measured in an ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). A total of 500 ng DNA was treated with sodium bisulfite e using the EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA, USA).
Amplicon library preparation
Given the low complexity of bisulfite-treated DNA and inherent limitations for PCR primer design (Li & Dahiya 2002), several alternative fusion PCR primers were designed and tested for each interrogated gene. Specifically amplified portions of the CpG islands that exhibited balanced amplification of unmethylated and methylated alleles were included in the amplicon library. A total of 13 and 25 CpG dinucleotides could be covered by the amplicons in SDHC and SDHD, respectively, adjoining the transcriptional start sites. The portions of promoter CpG islands with 14 and 15 CpG dinucleotides could be amplified for SDHA and SDHB respectively. PCR amplification was carried out with fusion primers with inner template-specific sequences (Table 2) in 50 μl reactions that contained 2.0 μl bisulfite-converted DNA, 1.5 mM MgCl2, 200 mM dNTP, 300 nM primers, and 0.52 units HotStar Taq DNA polymerase (Qiagen). An amplification program was used that was previously described with minor modification (Melki et al. 1999). Amplification of SDHA, SDHB, and SDHD loci was started by an initial activation of the HotStar Taq DNA polymerase at 95 °C for 15 min. The initial amplification cycle was denaturation at 95 °C for 1 min, annealing at 60 °C for 2 min, and elongation at 72 °C for 3 min. This procedure was continued for 20 cycles, reducing the annealing temperature by 0.5 °C each cycle, followed by 40 cycles of 1 min denaturation at 95 °C, 2 min annealing at 50 °C, and 2 min elongation at 72 °C. SDHC locus was amplified by employing a similar program with 61 °C as starting annealing temperature that was reduced by 0.5 °C each cycle for 20 cycles, followed by 40 cycles with annealing temperature of 51 °C. About 10 μl of each reaction was examined on 2.5% agarose gels.
Table 2.
Sequences of the PCR primers for amplicon library preparation and qPCR
| Primer ID | Primer sequences (5′–3′) | Amplicon length (bp)a | Number of CpGs quantified by massive parallel sequencing |
|---|---|---|---|
| Sequences of the template specific 3′-portion of the fusion PCR primersb | |||
| SDHA-F | AGTTATTTTAAGTTTGTTTATATGTGATTT | 236a | 14 |
| SDHA-R | ATAAACACCAACATTTTTAAAACCC | ||
| SDHB-F | TAGTTTGGTTAAGATGATGAAATTT | 302a | 15 |
| SDHB-R | CTTTCAAAAAATAAAACTAAAACTTAAATA | ||
| SDHC-F | GAAAATAATTAGTAAATTAGTTAGGTAG | 198a | 13 |
| SDHC-R | ACTAAAATCACCTCAACAACAAC | ||
| SDHD-F | TATTAAGGAAGGTGAAATTTTTTTT | 313a | 25 |
| SDHD-R | TCCTAAAAACTCAAAATCATCCAC | ||
| Sequences of the qPCR primers | |||
| SDHA-F | TGGGAACAAGAGGGCATCTG | 86 | |
| SDHA-R | CCACCACTGCATCAAATTCATG | ||
| SDHB-F | ACCTTCCGAAGATCATGCAG | 86 | |
| SDHB-R | CTTCGGGTGCAAGCTAGAGT | ||
| SDHC-F | TGGCACTGGTATTGCTTTGA | 78 | |
| SDHC-R | GACTCAAAGTTCCCAGGGAGT | ||
| SDHD-F | GGTGTGGAGTGCAGCACATA | 88 | |
| SDHD-R | ACAACCCTCTCGCTAGTCCA | ||
Template-specific portion of each amplicon is shown. Given additional auxiliary 35-mer sequences at both ends, real amplicon size is 70 bp longer for fusion PCVR primers.
The complete PCR primer sequence includes an additional 35-mer portion at 5′-end. It serves for binding to the DNA capture beads and annealing the emPCR amplification primers and the sequencing primer. In addition, it contains the sequencing tetranucleotide key and 10-mer multiple identifier (MID) for demultiplexing reads. A total of six MIDs (MID1-6) were used from the 454Standard MID set.
To control possible amplification bias, appropriate control DNA samples were included as described earlier (Moskalev et al. 2011). Fully methylated and unmethylated human control DNA that had been treated with bisulfite was obtained (EpiTect PCR control DNA; Qiagen) and mixed in different ratios to obtain samples that represent distinct methylation percentages of 0, 25, 50, 75, and 100% respectively. A total of 15 ng calibration DNA was used for the amplification of each locus.
Amplicons were purified by using the Agencourt AMPure XP Kit (Beckman Coulter, Beverly, MA, USA) and quantified by fluorometry in triplicates using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies) and Synergy 2 Multi-Mode Microplate Reader (Biotek, Winooski, VT, USA) as directed by the manufacturers. Finally, the library was pooled in equimolar ratios, and the concentration was adjusted to 107 molecules/μl.
Quantification of CpG dinucleotide methylation by massive parallel bisulfite sequencing
Massive parallel sequencing was carried out using the GS Junior Titanium chemistry, (Roche) as described earlier (Moskalev et al. 2013). All reads were processed and aligned to the bisulfite-modified human reference sequences of SDHA, SDHB, SDHC, and SDHD using the Amplicon Variant Analyser Software v. 2.5 from Roche. Quantification of CpG methylation was made using the BISMA Software (BISMA Software, Bremen, Germany; Rohde et al. 2010).
Quantitative PCR
Total RNA was isolated from snap-frozen tumor samples and non-neoplastic control specimens using the RNeasy Mini Kit (Qiagen). A total of 1 μg total RNA was reverse transcribed with the QuantiTect RT Kit (Qiagen), and relative mRNA expression levels of all four SDH subunits A, B, C, and D were measured in triplicates using the QuantiTect SYBR Green PCR Kit (Qiagen) and gene-specific primers (Table 2). The relative abundance of each subunit was calculated in relation to the mean abundance of all four subunits for each sample separately.
Western blotting
Protein lysates were isolated from snap-frozen tumor and non-neoplastic tissues, and 10–50 μg of lysate were loaded on a western blot. Protein levels were evaluated for SDHB (1:500 dilution, HPA002868, Sigma–Aldrich) and SDHC (1:1000 dilution, ab155999, Abcam, Cambridge, UK), with ACTB (1:5000 dilution, A5441, Sigma–Aldrich) serving as loading control.
Complex II/IV activity
Protein lysates were extracted from snap-frozen tumor tissues and GIST882 cell line. Enzyme activity of complexes II and IV were measured in triplicates using complex II (ab109908, Abcam) and complex IV (ab109909) Human Enzyme Activity Microplate Assay Kits respectively.
Results
The gene loci encoding for SDHB and SDHC subunits are aberrantly hypermethylated in tumors of CT patients
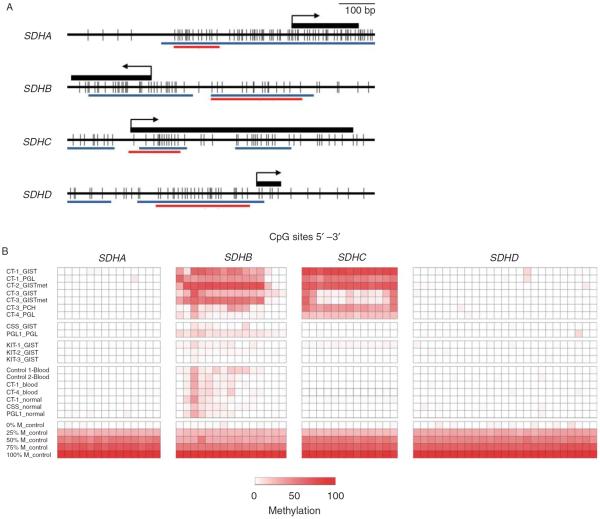
The genes coding for SDH subunits harbor extensive CpG islands in the promoter and first exonic regions (Fig. 2A). Because aberrant gains of DNA methylation in normally unmethylated CpG islands in vicinity of the transcriptional start sites frequently contributes to epigenetic silencing of cancer-relevant genes (Baylin & Jones 2011), we performed broad, high-resolution, and quantitative analysis of DNA methylation by means of massive parallel bisulfite sequencing within the genes' CpG islands in our set of samples. DNA methylation patterns were distinct for the four SDH subunits, and also comparing tumors associated with different conditions or genotypes. While DNA methylation was different for the gene loci encoding for the SDHB and SDHC subunits, there was no significant DNA methylation observed at the gene loci encoding for the SDHA and SDHD subunits in any of the tumors (Fig. 2B). Specific DNA methylation patterns were observed for the subunits SDHB and SDHC in the seven tumors of the four CT patients. SDHB was extensively methylated at 12 of 15 analyzed CpGs in the range of 10–77% in the GISTs and PGL of the first three CT patients, while the pulmonary chondroma of the third patient and the PGL of the fourth patient were only slightly methylated at few varying CpG dinucleotides (2–39%). SDHC was heavily methylated in the range of 16–80% at all 13 analyzed CpGs in the GISTs and PGLs of patients CT-1, CT-2, and CT-4, while unequal methylation load was observed in the GIST and the respective liver metastasis of patient CT-3 with only four CpGs sites being hypermethylated (14–64%). Interestingly, the pulmonary chondroma from the same patient, CT-3, was highly methylated at all 13 analyzed CpGs. Substantially lower (0–19%) DNA methylation degree was detected in the two tumors of the patients with CSS and PGL1 at few of the CpGs located in the proximity of SDHB, while SDHC was completely unmethylated at all 13 CpGs in the non-CT tumors (0–2%). Three KIT-mutated GISTs displayed no significant DNA methylation at any of the analyzed CpGs among all four SDH subunits A, B, C, and D. In the non-neoplastic controls from healthy donors (one male and female each) and patients with CT, CSS and PGL1, there was low DNA methylation at few varying CpG dinucleotides in the proximity of SDHB.
Figure 2.
High-resolution quantitative DNA methylation analysis of SDH subunits A, B, C and D. (A) CpG maps of the interrogated regions in the proximity of the gene loci encoding for the four SDH subunits. Individual CpG dinucleotides are represented by black vertical bars. The position of the first exon is shown as a black rectangle. The arrow indicates the transcriptional start sites. The red horizontal bars (denoted `454 SEQ') specify the CpG sites quantified by massive parallel bisulfite sequencing; the blue bars indicate CpG islands as analyzed by MethPrimer Software (MethPrimer Software, San Francisco, CA, USA). (B) DNA methylation patterns of the four SDH subunits in different tumor and control tissues. Each row represents a sample, and each column corresponds to an individual CpG dinucleotide. The degree of methylation was measured by massive parallel bisulfite sequencing and is color coded ranging from white (0%) to dark red (100%). Note the human control DNAs at the bottom representing distinct methylation percentages of 0, 25, 50, 75, and 100% respectively.
In summary, the gene locus of SDHC was specifically methylated at high levels in all tumors associated with CT, which was not observed in tumors associated with CSS or PGL1, sporadic GISTs or non-neoplastic controls. The gene locus of SDHB was also methylated at higher levels in tumors associated with CT compared with other tumors and non-neoplastic controls, although the pattern was less distinctive.
SDHC mRNA expression is downregulated and associated with the methylation status in tumors of CT patients
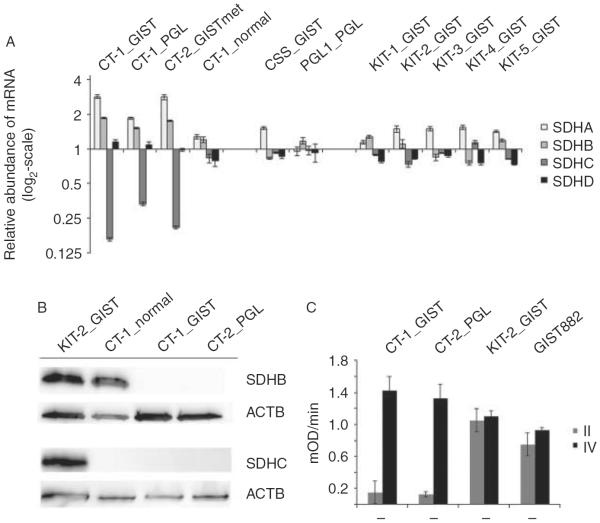
In order to determine the role of methylation in the regulation of expression, mRNA levels of SDH subunits A, B, C, and D were quantified using qPCR. SDHC was specifically downregulated in tumors associated with CT, while a balanced expression of all four subunits was observed in the non-CT tumor samples. Specifically, the qPCR analysis revealed a three- to sevenfold reduction in the relative abundance of SDHC mRNA compared with the other three subunits in particular in the tumor tissues of two GISTs and a PGL derived from two patients with CT (Fig. 3A). In contrast, the four subunits A, B, C, and D revealed a balanced expression with less than twofold differences of their relative abundance in tumor tissues of a GIST and a PGL in association with CSS and PGL1, respectively, as well as in sporadic GISTs with activating KIT mutations.
Figure 3.
Reduced mRNA expression and complete loss of SDHC protein confers to functional impairment of complex II activity in CT. (A) Quantitative mRNA expression analysis of all four SDH subunits A, B, C, and D reveals a three- to sevenfold reduction in SDHC mRNA abundance in CT tumors. The relative abundance for each SDH subunit was calculated in relation to the mean abundance of all four subunits within each individual tissue sample. (B) Complete loss of SDHC protein in the GIST and PGL tissues of CT-1 patient compared with KIT-mutated GIST. (C) Reducedactivity of complex II in the GIST and PGL tissues of CT-1 patient compared with KIT-mutated GIST and GIST882 cell line 1. Note the virtually equal activity of complex IV among all samples.
Loss of SDHC protein in tumors of CT patients corresponds to reduced complex II activity
Next, we assessed the abundance of SDHC using western blotting. Concordant with SDHC mRNA expression data, SDHC protein was lost in the GIST and the PGL of CT-1 patient, while it could be detected at high levels in a sporadic GIST with KIT mutation that was used as control (Fig. 3B). SDHB protein was similarly lost in the GIST and PGL tissue (Fig. 3B). There was no SDHC detectable by western blotting in the gastric mucosa of CT-1 patient, although qPCR revealed low but virtually equal mRNA expression of the four subunits in this tissue sample (Fig. 3A).
To confirm the functional effect of SDHC loss in the tumor tissues, the activity of complex II was determined. Accordingly, the activity of complex II was fivefold reduced in the GIST and PGL tissue of patient CT-1 compared with tissue from a sporadic KIT-mutated GIST or the GIST882 cell line (Fig. 3C). Activity of complex IV was used as a control for equal protein input, and was virtually the same in the CT tumors and the controls.
Discussion
Despite a similar morphology including loss of SDHB immunostaining, no germline or somatic mutations within any of the four complex II subunits SDHA, SDHB, SDHC, and SDHD have been found in CT patients and tumors compared with patients and tumors with CSS or familial PGL (Matyakhina et al. 2007, McWhinney et al. 2007, Pasini et al. 2008). In the current study, using broad and high-resolution quantification of DNA methylation by means of massive parallel bisulfite sequencing, we observed a DNA methylation signature within the CpG island in proximity of SDHC that was specific for GISTs/PGLs associated with CT, and absent in GISTs/PGLs associated with SDH germline mutations, or sporadic GISTs with activating KIT mutations. This dense DNA hypermethylation was correlated with reduced SDHC mRNA expression and a loss of SDHC protein in GIST/PGL tissues from CT patients, compared with a balanced mRNA expression of the four subunits as well as retained SDHC protein in the control tumors. Collectively, these data suggest for the first time that epigenetic inactivation of the SDHC gene locus with consequent reduction or loss of SDHC at mRNA and protein levels is a plausible cause for tumor development in patients with CT.
The SDHC gene encodes a hydrophobic protein that anchors the SDH complex to the mitochondrial inner membrane (Pasini & Stratakis 2009, Welander et al. 2011). Germline mutations in SDHC have been identified as a cause of familial PGL3 syndrome (Niemann & Müller 2000). PGL3 follows an autosomal-dominant inheritance pattern with germline mutation in one allele and additional somatic loss of the second allele in the tumor, resulting in a loss of SDHC protein with functional impairment of the SDH complex enzyme activity (Niemann & Müller 2000). Overall, SDHC-germline mutation carriers are much less frequent compared with patients with germline SDHD or SDHB mutations among cohorts of patients with PGLs/pheochromocytomas (Schiavi et al. 2005, Mannelli et al. 2009, Lefebvre et al. 2012). Up to now, 47 unique SDHC mutations from 42 individuals are included in the Leiden Open Variation Database (Bayley et al. 2005). Notably, the clinical phenotypes are distinct among patients with different familial PGL syndromes. Patients with SDHC germline mutations appear to be significantly older with a mean age of 38 years at first presentation compared with patients with SDHB or SDHD germline mutations (Schiavi et al. 2005, Mannelli et al. 2009, Lefebvre et al. 2012). While a first report on SDHC-germline mutation carriers observed only PGLs of the head and neck that were only rarely multiple and virtually exclusively benign (Schiavi et al. 2005), later studies identified SDHC-related adrenal medullary PGLs (pheochromocytomas) as well (Peczkowska et al. 2008), and mediastinal PGL was identified as the second most common involved anatomical localization (Else et al. 2014). Moreover, malignant PGLs have also been described in association with SDHC germline mutations (Bickmann et al. 2014), and SDHC germline mutations were found in apparently sporadic PGLs (Bayley et al. 2006). Interestingly, germline mutations in SDHC are also present in CSS, a condition with autosomal-dominant inheritance pattern and syndromal occurrence of PGLs and GISTs in affected patients (Carney & Stratakis 2002). Similar to the familial PGL syndromes, germline mutations of the other subunits of the SDH complex can also cause CSS. Interestingly, SDHC mutations were relatively frequent in the first description of CSS, occurring in two of seven families with SDH germline mutations (McWhinney et al. 2007, Pasini et al. 2008). Notably, both families with CSS in relation to SDHC germline mutations presented with GISTs as the first tumor, and other families with SDHC germline mutation and syndromal occurrence of GISTs and PGLs have been described as well (Stratakis & Carney 2009, Gill et al. 2013). In summary, patients with SDHC germline mutations have a high risk to develop PGLs, GISTs, and pheochromocytomas, representing a clinical phenotype with significant overlap to patients with CT (Pasini & Stratakis 2009, Welander et al. 2011). Thus, epigenetic silencing of the SDHC gene locus with concurrent functional loss on the mRNA and protein levels as suggested in the current study is a reasonable alternate mechanism for the development of CT, explaining the closely related phenotype compared with SDHC-germline mutation carriers.
Both GISTs (Killian et al. 2013) and PGLs (Letouzé et al. 2013) with SDH mutations have recently been found to display a hypermethylator phenotype, which is distinct compared with PGLs and GISTs with different genetic backgrounds (e.g. NF1 mutations and KIT mutations). These studies link the metabolic SDH complex to DNA methylation, most likely through accumulation of succinate with consecutive inhibition of 2-oxoglutarate-dependent oxidative demethylation. In contrast to the broad global hypermethylation pattern observed on the DNA level in SDH-deficient PGLs, only a subset of 191 genes displayed a correlated downregulation of mRNA expression as determined by integrative methylation and gene expression analysis (Letouzé et al. 2013). Interestingly, no correlation between SDHC methylation and mRNA expression has been reported in that study focusing on familial PGLs with SDH germline mutations (Letouzé et al. 2013). In contrast, in the current study analyzing CT tumors without SDH mutations, aberrant DNA methylation of SDHC was correlated with a reduction of SDHC mRNA expression and a loss of SDHC protein. This pattern was not found in the GIST and PGL associated with SDH-deficiency in the context of SDHB and SDHD germline mutations. Accordingly, we consider the aberrant DNA methylation pattern of SDHC specific for tumors in association with CT, and distinct from the global DNA hypermethylation pattern generally observed in tumors with SDH deficiency according to SDH germline mutations (Killian et al. 2013, Letouzé et al. 2013). While no methylation of SDHA and SDHD was observed in any of the analyzed tumors in the current study, SDHB was hypermethylated in some of the cases. In general, SDHB methylation was higher in tumors from CT patients compared with the tumors of patients with SDH germline mutations or sporadic KIT-mutated GISTs. However, there were two tumors associated with CT that showed similar levels compared with the SDH-mutated tumors, and there was a balanced SDHB mRNA expression both in tumors with SDH mutations and from CT patients. On the protein level, tumors from CT patients as well as tumors with SDH mutations displayed a complete loss of SDHB as determined by immunohistochemistry. This phenomenon of a general loss of SDHB subunit on the protein level irrespective of the mutated subunit of the SDH complex is well known (van Nederveen et al. 2009), and probably related to a posttranscriptional HIF1α-dependent feedback loop (Dahia et al. 2005). In contrast, no such effect has been reported for SDHC so far. Thus, we consider the aberrant DNA methylation of SDHC with concurrent reduction in SDHC mRNA expression and complete loss of SDHC in the tumor tissue a specific and tumor-initiating event, which is distinct from the global DNA hypermethylation patterns that have been recently described in tumors with SDH deficiency (Killian et al. 2013, Letouzé et al. 2013).
Apart from the lack of both germline and somatic mutations among SDH subunits or other genes found to be mutated in familial or sporadic PGLs and GISTs, little is known about the genetic changes within CT patients and tumors. One comprehensive study analyzed chromosomal imbalances in 41 tumor samples derived from 37 CT patients (Matyakhina et al. 2007). Of note, the most frequent and greatest contiguous change involved loss of the chromosomal region 1q12–q23.3, thus including the SDHC gene locus located at 1q23.3. The second most common change was alterations of the short arm of chromosome 1, corresponding to the SDHB gene locus located at 1p36.1–p35. This study strongly supports our current finding of concurrent hypermethylation of the SDHC gene locus and loss of SDHC expression, in that somatic genetic loss of the SDHC gene locus as reported in the study by Matyakhina et al. (2007) will further enhance the functional loss of SDHC. To date, no other genetic event has been reported in CT patients or tumors. Regarding the syndromal character of CT with strong predilection for the female gender in contrast to the lack of familial cases or inheritance (Carney et al. 1977, Carney 2009), future studies of this fascinating disease employing genome-wide DNA sequencing are needed to identify or exclude recurrent genetic aberrations beyond inactivation of the SDHC gene locus.
Acknowledgements
The authors thank Simone Hebele (Diagnostic Molecular Pathology, Institute of Pathology, University Hospital, Friedrich-Alexander University Erlangen-Nuremberg, Erlangen, Germany) for excellent technical assistance.
Funding E A Moskalev is supported by a research grant from the Interdisciplinary Centre for Clinical Research (IZKF) at the University of Erlangen-Nuremberg. I-M Schaefer is supported by a research grant from the Dr Mildred Scheel Stiftung für Krebsforschung (no. 110822).
Footnotes
Declaration of interest The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Author contribution statement F Haller, E A Moskalev, C A Stratakis, and A Agaimy contributed equally to this work.
References
- Bayley JP, Devilee P, Taschner PE. The SDH mutation database: an online resource for succinate dehydrogenase sequence variants involved in pheochromocytoma, paraganglioma and mitochondrial complex II deficiency. BMC Medical Genetics. 2005;6:39. doi: 10.1186/1471-2350-6-39. doi:10.1186/1471-2350-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley JP, van Minderhout I, Weiss MM, Jansen JC, Oomen PH, Menko FH, Pasini B, Ferrando B, Wong N, Alpert LC, et al. Mutation analysis of SDHB and SDHC: novel germline mutations in sporadic head and neck paraganglioma and familial paraganglioma and/or pheochromocytoma. BMC Medical Genetics. 2006;7:1. doi: 10.1186/1471-2350-7-1. doi:10.1186/1471-2350-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome – biological and translational implications. Nature Reviews. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. doi:10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmann JK, Sollfrank S, Schad A, Musholt TJ, Springer E, Miederer M, Bartsch O, Papaspyrou K, Koutsimpelas D, Mann WJ, et al. Phenotypic variability and risk of malignancy in SDHC-linked paragangliomas: lessons from 3 unrelated cases with an identical germline mutation (p.Arg133*) Journal of Clinical Endocrinology and Metabolism. 2014;99:E489–E496. doi: 10.1210/jc.2013-3486. doi:10.1210/jc.2013-3486. [DOI] [PubMed] [Google Scholar]
- Carney JA. Carney triad: a syndrome featuring paraganglionic, adrenocortical, and possibly other endocrine tumors. Journal of Clinical Endocrinology and Metabolism. 2009;94:3656–3662. doi: 10.1210/jc.2009-1156. doi:10.1210/jc.2009-1156. [DOI] [PubMed] [Google Scholar]
- Carney JA, Stratakis CA. Familial paraganglioma and gastric stromal sarcoma: a new syndrome distinct from the Carney triad. American Journal of Medical Genetics. 2002;108:132–139. doi: 10.1002/ajmg.10235. doi:10.1002/ajmg.10235. [DOI] [PubMed] [Google Scholar]
- Carney JA, Sheps SG, Go VL, Gordon H. The triad of gastric leiomyosarcoma, functioning extra-adrenal paraganglioma and pulmonary chondroma. New England Journal of Medicine. 1977;296:1517–1518. doi: 10.1056/NEJM197706302962609. doi:10.1056/NEJM197706302962609. [DOI] [PubMed] [Google Scholar]
- Dahia PL, Ross KN, Wright ME, Hayashida CY, Santagata S, Barontini M, Kung AL, Sanso G, Powers JF, Tischler AS, et al. A HIF1α regulatory loop links hypoxia and mitochondrial signals in pheochromocytomas. PLoS Genetics. 2005;1:72–80. doi: 10.1371/journal.pgen.0010008. doi:10.1371/journal.pgen.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else T, Marvin ML, Everett JN, Gruber SB, Arts HA, Stoffel EM, Auchus RJ, Raymond VM. The clinical phenotype of SDHC-associated hereditary paraganglioma syndrome (PGL3) Journal of Clinical Endocrinology and Metabolism. 2014 doi: 10.1210/jc.2013-3853. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill AJ, Lipton L, Taylor J, Benn DE, Richardson AL, Frydenberg M, Shapiro J, Clifton-Bligh RJ, Chow CW, Bogwitz M. Germline SDHC mutation presenting as recurrent SDH deficient GIST and renal carcinoma. Pathology. 2013;45:689–691. doi: 10.1097/PAT.0000000000000018. doi:10.1097/PAT.0000000000000018. [DOI] [PubMed] [Google Scholar]
- Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI, Jr, Jahromi MS, Xekouki P, et al. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discovery. 2013;3:648–657. doi: 10.1158/2159-8290.CD-13-0092. doi:10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Borson-Chazot F, Boutry-Kryza N, Wion N, Schillo F, Peix JL, Brunaud L, Finat A, Calender A, Giraud S. Screening of mutations in genes that predispose to hereditary paragangliomas and pheochromocytomas. Hormone and Metabolic Research. 2012;44:334–338. doi: 10.1055/s-0032-1306308. doi:10.1055/s-0032-1306308. [DOI] [PubMed] [Google Scholar]
- Letouzé E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. doi:10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. doi:10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- Mannelli M, Castellano M, Schiavi F, Filetti S, Giacchè M, Mori L, Pignataro V, Bernini G, Giachè V, Bacca A, et al. Clinically guided genetic screening in a large cohort of Italian patients with pheochromocytomas and/or functional or nonfunctional paragangliomas. Journal of Clinical Endocrinology and Metabolism. 2009;94:1541–1547. doi: 10.1210/jc.2008-2419. doi:10.1210/jc.2008-2419. [DOI] [PubMed] [Google Scholar]
- Matyakhina L, Bei TA, McWhinney SR, Pasini B, Cameron S, Gunawan B, Stergiopoulos SG, Boikos S, Muchow M, Dutra A, et al. Genetics of Carney triad: recurrent losses at chromosome 1 but lack of germline mutations in genes associated with paragangliomas and gastrointestinal stromal tumors. Journal of Clinical Endocrinology and Metabolism. 2007;92:2938–2943. doi: 10.1210/jc.2007-0797. doi:10.1210/jc.2007-0797. [DOI] [PubMed] [Google Scholar]
- McWhinney SR, Pasini B, Stratakis CA. Familial gastrointestinal stromal tumors and germ-line mutations. New England Journal of Medicine. 2007;357:1054–1056. doi: 10.1056/NEJMc071191. doi:10.1056/NEJMc071191. [DOI] [PubMed] [Google Scholar]
- Melki JR, Vincent PC, Clark SJ. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Research. 1999;59:3730–3740. [PubMed] [Google Scholar]
- Merkelbach-Bruse S, Dietmaier W, Füzesi L, Gaumann A, Haller F, Kitz J, Krohn A, Mechtersheimer G, Penzel R, Schildhaus HU, et al. Pitfalls in mutational testing and reporting of common KIT and PDGFRA mutations in gastrointestinal stromal tumors. BMC Medical Genetics. 2010;11:106. doi: 10.1186/1471-2350-11-106. doi:10.1186/1471-2350-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalev EA, Zavgorodnij MG, Majorova SP, Vorobjev IA, Jandaghi P, Bure IV, Hoheisel JD. Correction of PCR-bias in quantitative DNA methylation studies by means of cubic polynomial regression. Nucleic Acids Research. 2011;39:e77. doi: 10.1093/nar/gkr213. doi:10.1093/nar/gkr213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalev EA, Stöhr R, Rieker R, Hebele S, Fuchs F, Sirbu H, Mastitsky SE, Boltze C, König H, Agaimy A, et al. Increased detection rates of EGFR and KRAS mutations in NSCLC specimens with low tumour cell content by 454 deep sequencing. Virchows Archiv. 2013;462:409–419. doi: 10.1007/s00428-013-1376-6. doi:10.1007/s00428-013-1376-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nederveen FH, Gaal J, Favier J, Korpershoek E, Oldenburg RA, de Bruyn EM, Sleddens HF, Derkx P, Rivière J, Dannenberg H, et al. An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. Lancet Oncology. 2009;10:764–771. doi: 10.1016/S1470-2045(09)70164-0. doi:10.1016/S1470-2045(09)70164-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann S, Müller U. Mutations in SDHC cause autosomal dominant paraganglioma, type 3. Nature Genetics. 2000;26:268–270. doi: 10.1038/81551. doi:10.1038/81551. [DOI] [PubMed] [Google Scholar]
- Pasini B, Stratakis CA. SDH mutations in tumorigenesis and inherited endocrine tumours: lesson from the phaeochromocytoma–paraganglioma syndromes. Journal of Internal Medicine. 2009;266:19–42. doi: 10.1111/j.1365-2796.2009.02111.x. doi:10.1111/j.1365-2796.2009.02111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini B, McWhinney SR, Bei T, Matyakhina L, Stergiopoulos S, Muchow M, Boikos SA, Ferrando B, Pacak K, Assie G, et al. Clinical and molecular genetics of patients with the Carney–Stratakis syndrome and germline mutations of the genes coding for the succinate dehydrogenase subunits SDHB, SDHC, and SDHD. European Journal of Human Genetics. 2008;16:79–88. doi: 10.1038/sj.ejhg.5201904. doi:10.1038/sj.ejhg.5201904. [DOI] [PubMed] [Google Scholar]
- Peczkowska M, Cascon A, Prejbisz A, Kubaszek A, Cwikła BJ, Furmanek M, Erlic Z, Eng C, Januszewicz A, Neumann HP. Extra-adrenal and adrenal pheochromocytomas associated with a germline SDHC mutation. Nature Clinical Practice. Endocrinology & Metabolism. 2008;4:111–115. doi: 10.1038/ncpendmet0726. doi:10.1038/ncpendmet0726. [DOI] [PubMed] [Google Scholar]
- Rattenberry E, Vialard L, Yeung A, Bair H, McKay K, Jafri M, Canham N, Cole TR, Denes J, Hodgson SV, et al. A comprehensive next generation sequencing-based genetic testing strategy to improve diagnosis of inherited pheochromocytoma and paraganglioma. Journal of Clinical Endocrinology and Metabolism. 2013;98:1248–1256. doi: 10.1210/jc.2013-1319. doi:10.1210/jc.2013-1319. [DOI] [PubMed] [Google Scholar]
- Rohde C, Zhang Y, Reinhardt R, Jeltsch A. BISMA – fast and accurate bisulfite sequencing data analysis of individual clones from unique and repetitive sequences. BMC Bioinformatics. 2010;11:230. doi: 10.1186/1471-2105-11-230. doi:10.1186/1471-2105-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavi F, Boedeker CC, Bausch B, Peçzkowska M, Gomez CF, Strassburg T, Pawlu C, Buchta M, Salzmann M, Hoffmann MM, et al. Predictors and prevalence of paraganglioma syndrome associated with mutations of the SDHC gene. Journal of the American Medical Association. 2005;294:2057–2063. doi: 10.1001/jama.294.16.2057. doi:10.1001/jama.294.16.2057. [DOI] [PubMed] [Google Scholar]
- Stratakis CA, Carney JA. The triad of paragangliomas, gastric stromal tumours and pulmonary chondromas (Carney triad), and the dyad of paragangliomas and gastric stromal sarcomas (Carney–Stratakis syndrome): molecular genetics and clinical implications. Journal of Internal Medicine. 2009;266:43–52. doi: 10.1111/j.1365-2796.2009.02110.x. doi:10.1111/j.1365-2796.2009.02110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander J, Söderkvist P, Gimm O. Genetics and clinical characteristics of hereditary pheochromocytomas and paragangliomas. Endocrine-Related Cancer. 2011;18:253–276. doi: 10.1530/ERC-11-0170. doi:10.1530/ERC-11-0170. [DOI] [PubMed] [Google Scholar]