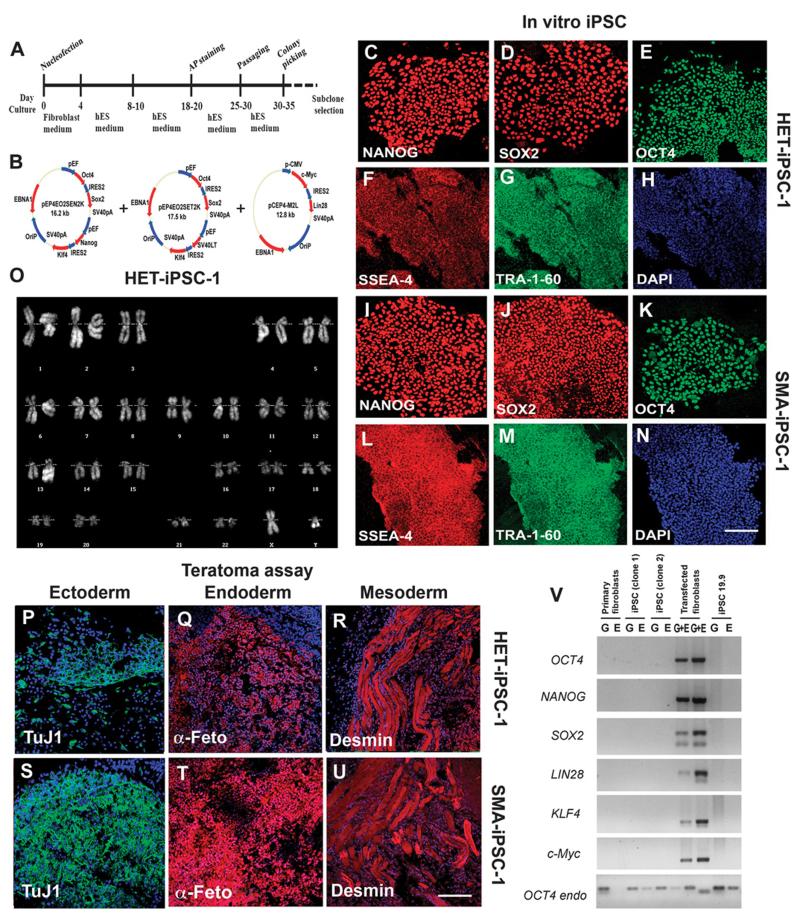
Fig. 1.
Reprogramming human fibroblasts of an SMA patient and his father without genomic vector integration, and selection of iPSC clones. (A) Schematic representation of the nonviral reprogramming protocol for adult human fibroblasts. (B) Episomal vector maps. (C to N) Immunocytochemical characterization of iPSC clones derived from an SMA patient (SMA-iPSCs) and his heterozygous father (HET-iPSCs). These iPSCs express pluripotency transcription factors including NANOG (red), SOX2 (red), and OCT4 (green) (C to E and I to K), as well as stem cell surface markers (SSEA-4, red) and TRA-1-60 (green). Blue, 4′,6-diamidino-2-phenylindole (DAPI) nuclear stain. (F) to (H) and (L) to (N) are representative images. (O) These cells (HET-iPSCs) were karyotypically normal. (P to U) SMA-iPSC clones and HET-iPSCs form teratomas in vivo that contain the three germ layers as shown by representative images from one SMA-iPSC and one HET-iPSC clone: ectoderm (TuJ1, green), mesoderm (desmin, red), and endoderm (α-fetoprotein, red). (V) PCR analysis of episomal DNA in iPSC clones. Genomic (G) and episomal (E) DNAs from nontransfected and vector-transfected adult human fibroblasts were used as negative (–) and positive (+) controls, respectively. No plasmid integration into iPSCs was observed. Scale bars, 100 μm (C to N); 150 μm (L to N) and (P to U).