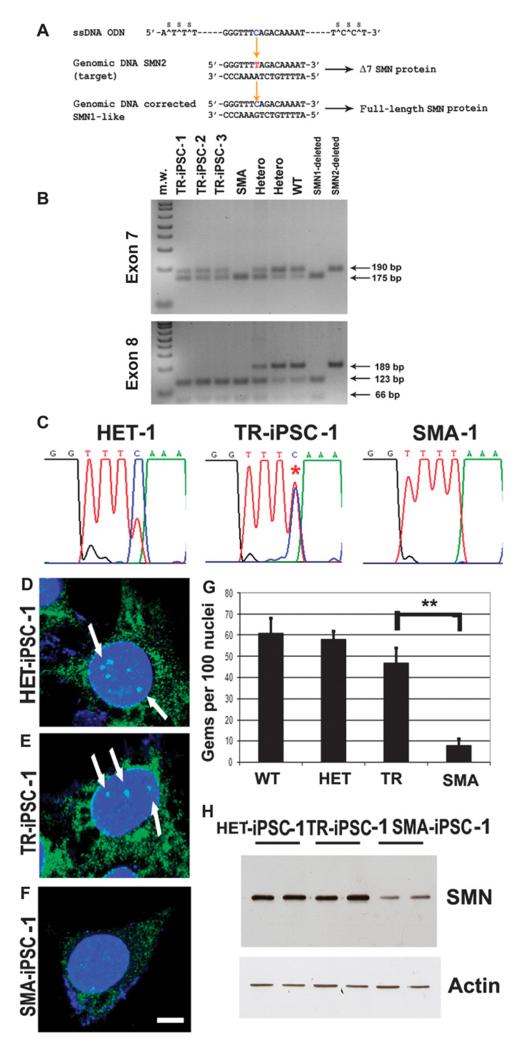
Fig. 2.
Genetic correction by modification of the SMN2 gene using oligonucleotides in SMA-iPSCs. (A) Targeted SMN2sequence and correcting oligonucleotides. The blue base in the oligonucleotides is the base that directs the targeting to the red base in the SMN2 sequence, which will render exon 7 in SMN2 like exon 7 in SMN1, resulting in its inclusion during splicing. (B and C) Restriction digest and cycle sequencing confirm base conversion in SMN2 in SMA-iPSCs. (B) The PCR products were subjected to restriction enzyme treatment: Dra1 for exon 7 (upper panel) and Dde1 for exon 8 (lower panel) (the enzymes Dra I and Dde I cleave the PCR products from SMN2 exons 7 and 8, respectively). In the corrected SMA-iPSCs, the presence of an uncut band corresponds to corrected exon 7, and the complete digested band (cut) corresponds to exon 8. This pattern corresponds to the modification of theSMN2 gene into the SMN1-like gene and rules out the possibility of a contamination with wild-type (WT) or heterozygous DNA. PCR electrophoresis showed the predicted band sizes: 175 bp for SMN1 and 190 bp forSMN2 for exon 7 (upper panel). The three SMA-iPSC clones showed the presence of both bands, supporting the exon 7 base conversion to that of SMN1. (C) The results of the sequencing reaction confirm the conversion. The asterisk on a double peak indicates that both T and C have been detected, implying that this sample is heterozygous for SMN1 and SMN2. (D to G) The number of gems detected by SMN immunocytochemistry was higher in the HET-iPSCs (HET), WT iPSCs, and oligonucleotide-treated SMA-iPSCs (TR) when compared to untreated SMA-iPSCs (SMA) (**P ≤ 0.01). Mean ± SD, n = 5 independent experiments per condition [one-way analysis of variance (ANOVA) followed by a Tukey’s post hoc test]. (H) Western blot analysis showed that corrected SMA-iPSCs had higher SMN protein concentrations than untreated SMA-iPSCs.