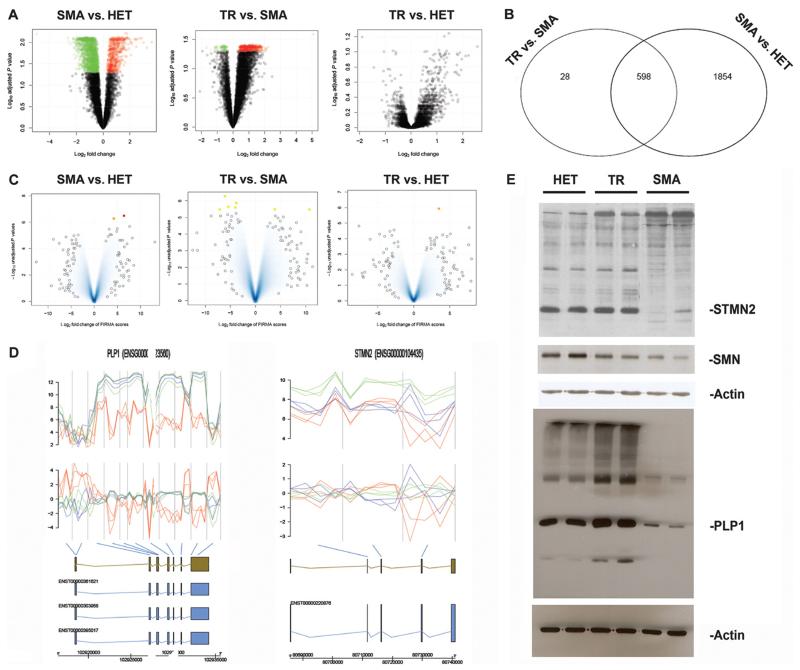
Fig. 4.
Global gene expression and splicing analysis of iPSC-derived motor neurons. (A) Volcano plot for the class comparisons of gene expression. In the upper region of the picture are the most deregulated genes (green, down-regulated; red, up-regulated). (B) Venn diagram comparing the common differentially expressed genes. (C) Result for the three classes from the splicing analysis. The most significant data are marked in red, orange, and yellow and listed in the Supplementary Materials. (D) Splicing analysis of some of the most significantly deregulated genes. Figures illustrate the genes found to be differentially spliced, indicating from top to bottom the expression intensities and FIRMA scores for the individual probes in the probe sets [untreated (red) and corrected (blue) SMA-iPSC motor neurons; green, HET-iPSC motor neurons]. The gene, different transcripts, and exons are illustrated, with the blue bars indicating the association of the probe sets with the individual exons. The most significant probe set based on the FIRMA score is highlighted. (E) Western blot analysis of the deregulated proteins in motor neurons from untreated iPSCs (SMA), corrected iPSCs (TR), and HET-iPSCs. There is down-regulation of SMN in motor neurons from untreated iPSCs (SMA) compared to those from corrected iPSCs (TR) and HET-iPSCs. The most commonSTMN2 isoform as well as PLP1 appear to be down-regulated in the motor neurons from untreated iPSCs (SMA) compared to those from corrected iPSCs (TR) and HET-iPSCs. In the same Western blot, there are also some other differentially expressed bands between the two samples.