Fig. 2.
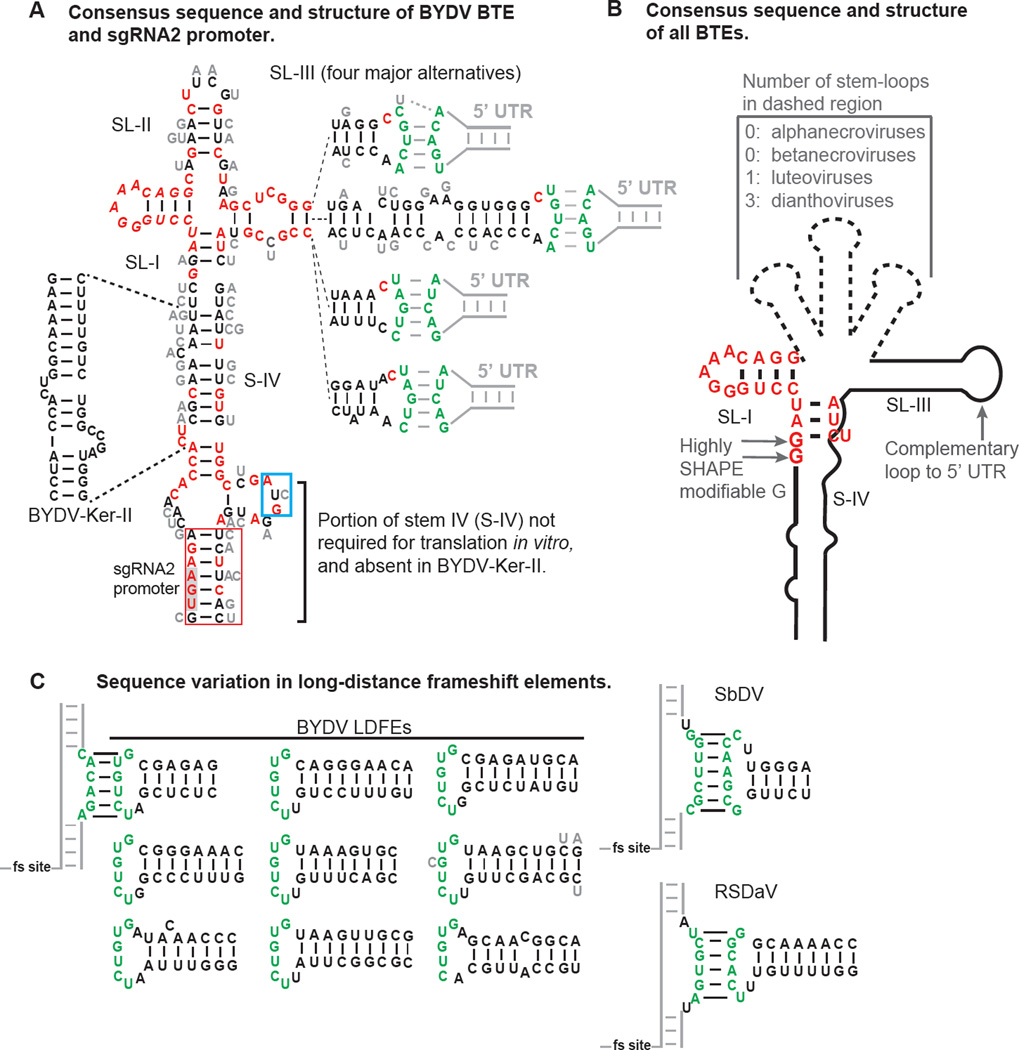
Secondary structures that regulate luteovirus translation. A. Secondary structure and base variations of BYDV BTEs. The 5’ end of the sequence shown is the 5’ end of sgRNA2. Sequence variation in fifty-one isolates of BYDV species is indicated, with invariant bases in red. Alternative bases found at each position are shown in gray beside each base. There are four major forms of the distal end of stem-loop III (SL-III), with each indicated separately, connected by dashed lines to the proximal end of SL-III. Green bases form kissing stem-loop base pairing with the stem-loop in the 5’ UTR indicated by gray lines. The sequence in the red box is sufficient for production of sgRNA2 by BYDV-PAV6. UGA (gray shading) is stop codon of ORF 5. AUG in blue box is the ORF 6 start codon. B. Consensus structure of known BTEs, which includes all viruses in the Luteovirus, Dianthovirus and Necrovirus genera. Note that the RCNMV BTE has no known loop complementary to the 5’ UTR. The first or second guanosine residue (but not both) in the 17 nt conserved sequence is highly modified by SHAPE probing in BTEs of all genera, indicating that it is not base paired and may protrude from the structure in an unusual way (Kraft et al., 2013). (Two of the fifty-one isolates contain an A at position 2 of the 17 nt conserved sequence. Their translation efficiency is unknown.) C. Base pairing of the long-distance frameshift elements (LDFE) in the 3’ UTR of luteoviruses with a bulge adjacent to frameshift site. Sequences found in 51 isolates of five BYDV species contain one of the nine structures shown. Note the lack of sequence conservation in the stem, but conservation of the loop that base pairs to the bulged helix, CACAGA, which is invariant in all 51 isolates adjacent to the frameshift site (fs site). One isolate (gray C, Genbank accession no. EU332331) has a mismatch, and another (BYDV-Ker-II, accession no. NC_021481, bottom right) has only five bases of complementarity. Different sequences participate in long-distance base pairing in SbDV and RSDaV. No obvious LDFE was identified in the 3’ UTR of BLRV.