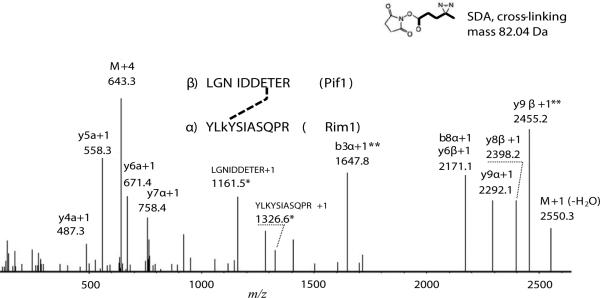
Figure 2. Identification of Pif1-Rim1 interaction site using SDA cross-linking.
The charge-deconvoluted tandem MS/MS spectrum obtained using HCD fragmentation from the precursor at 643.078 m/z is shown. Mass deviation from the theoretical mass was 4.83 ppm. The precursor retention time was 29 min, which is close to the expected value, based on the hydrophobicity of the cross-linked peptides. Mass deviation of the fragments was within 20 ppm, on average. Possibility of one C13 atom was allowed in calculations of the mass deviation. The most intense ions and the ions indicative of cross-link fragmentation were annotated by StavroX, xComb, and manually. Fragments contacting cross-links are shown in bold. Fragments used to identify the cross-linked residue on Rim1 are marked with (**). For the details of StavroX and xComb annotations see Supplemental Figure 4. Stavro score was 109, with the 5% false discovery rate corresponding to the score above 68, according to the Stavro decoy analysis (Supplemental Figure 4, C). Greek letters α and β correspond to the cross-linked peptides from Rim1 and Pif1, respectively. Fragments related to the loss of the cross-link are marked with asterisk (*). Letter ‘M’ notes precursor-related ions