Figure 1.
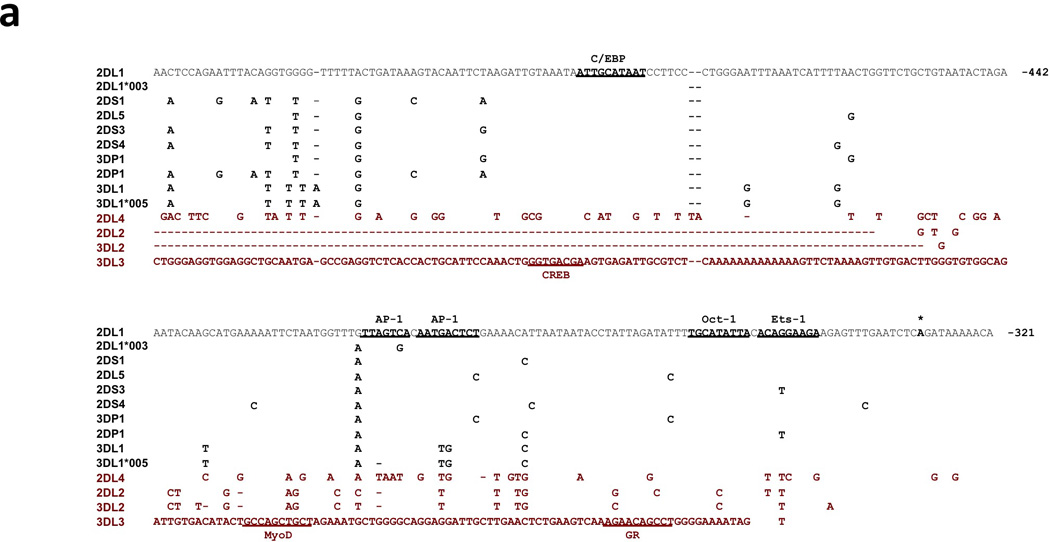
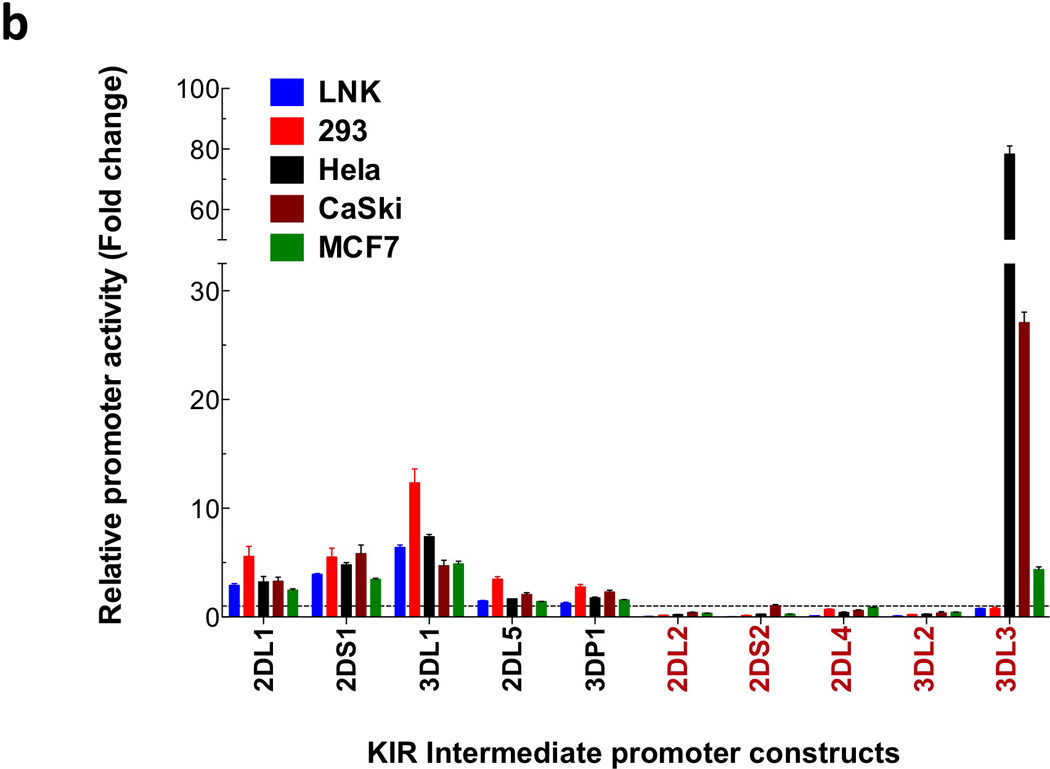
Comparison of the KIR2DL1 ProI region with other KIR genes. (a) The 240 bp ProI region of KIR2DL1 is shown, with only nucleotide differences displayed for other KIR genes. The location of the KIR2DL1 sequence relative to the start codon of the gene is indicated in bold at the right end of each line of sequence. Dashes indicate deletions relative to the KIR2DL1 sequence. Underlined bold sequence represents putative transcription factor-binding elements. The single bold A residue denoted by an asterisk indicates the transcription start site determined for KIR2DL1 ProI. KIR2DL1*003 represents the unique sequence found in the KIR2DL1*003 allele, and KIR3DL1*005 represent the sequence found in the KIR3DL1*001, *004, and *005 alleles. KIR listed in red print are genes with low homology to the KIR2DL1 ProI region. The complete sequence of the KIR3DL3 gene upstream of the Ets-1 site is shown due to a very low level of homology with KIR2DL1 in this region. (b) Luciferase activity of pGL3 constructs containing the KIR promoter regions depicted in (a). Constructs were transfected into the cell lines listed, and relative luciferase activity was determined 48 hours post-transfection. Values represent the mean, and error bars indicate the standard deviation of at least 3 independent experiments. KIR genes listed in red type correspond to the divergent KIR gene sequences identified in (a).
