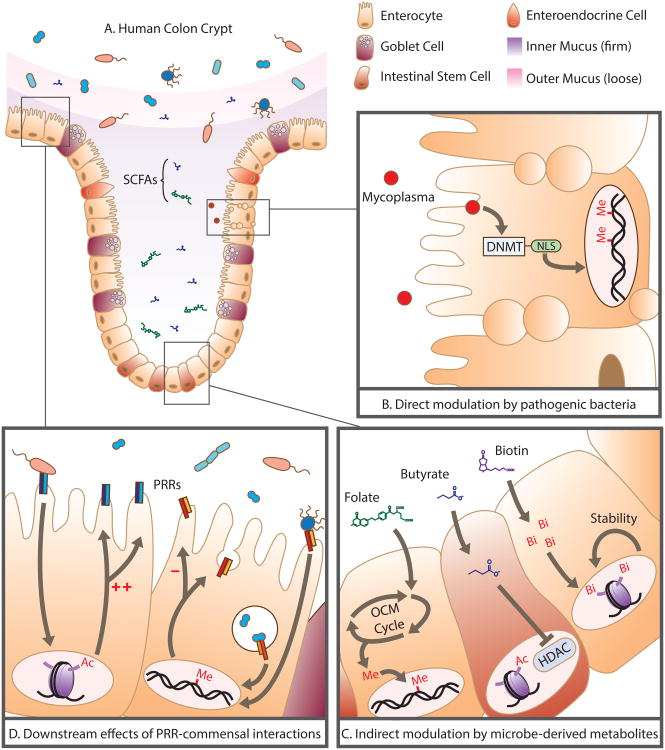
Figure 2.
Direct and indirect modulation of the human epigenome by pathogenic and commensal microbiota: (A) Human colonic physiology and its spatial relationship with the microbiome and the short chain fatty acids (SCFAs) and metabolites it produces. (B) Mycoplasma, an intracellular microbial pathogen, synthesizes DNA methyltransferases (DNMT) complete with nuclear localization signals (NLS) that penetrate the host nucleus and result in de novo methylation. (C) Microbe-derived metabolites (folate, butyrate, and biotin) indirectly modulate the host epigenome: Folate enters the one carbon metabolism cycle (OCM Cycle) to affect the bioavailability of methyl groups, butyrate acts as a potent histone deacetylase (HDAC) inhibitor, biotin availability for biotinylation promotes chromosomal stability. (D) Commensal interactions with pattern recognition receptors (PRRs) cause downstream transcriptional changes, mediated primarily through methylation and acetylation, resulting in increased/decreased production of PRRs as well as extra-epithelial responses.