Abstract
We have previously shown that complement component 3 (C3) is secreted by malignant epithelial cells. To understand the mechanism of upregulation of C3 expression in tumor cells we studied the C3 promoter; and identified that TWIST1 binds to the C3 promoter and enhances its expression. Because TWIST1 mediates epithelial-mesenchymal transition (EMT), we studied the effect of C3 on EMT and found that C3 decreased E-cadherin expression on cancer cells and promoted EMT. We showed that C3-induced reduction in E-cadherin expression in ovarian cancer cells was mediated by C3a and is Krüppel-like factor 5 (KLF5)-dependent. We investigated the association between TWIST1 and C3 in malignant tumors and in murine embryos. TWIST1 and C3 co-localized at the invasive tumor edges, and in the neural crest and limb buds of mouse embryos. Our results identified TWIST1 as a transcription factor that regulates C3 expression during pathologic and physiologic EMT.
Introduction
The complement system comprises several soluble and cell surface proteins, and is an important part of innate immunity. Activation of the complement system via classic, alternative, and lectin pathways converge on generation of C3 convertase that cleaves C3; and culminate in tagging of the complement-targeted cells by C3 degradation products, in generating anaphylatoxins (C3a and C5a), and in assembling membrane attack complexes (MAC; C5b-9) on the target membranes. In addition to fighting invading microorganisms, complement plays an important role in other physiologic and pathologic processes, including organogenesis, embryogenesis, and regulation of adaptive immunity (1,2). Complement also plays a role in the cell-to-cell communication, angiogenesis, organ regeneration, and cell migration (3).
C3 is a plasma protein that is synthesized in the liver, but endothelial cells, white blood cells, and epithelial cells also secrete complement proteins (4-7). We have found that malignant ovarian epithelial cells synthesize and secrete C3 (8), which in turn results in complement activation in the tumor microenvironment and increases cancer cell proliferation, invasion, and migration. However, the regulation of C3 expression in malignant cells is not well understood.
Previous studies have shown that Ccaat-enhancer-binding protein δ (C/EBP δ) regulates C3 in the liver (9) and in astrocytes and neurons (10); and vitamin D regulates C3 in osteoblasts (11). Here we report that TWIST1 (twist basic helix-loop-helix transcription factor 1) regulates expression of C3 in ovarian cancer cells. TWIST1 is an important transcription factor in epithelial-mesenchymal transition (EMT), which occurs in both physiologic (embryogenesis) (12,13) and pathologic (metastasis) conditions (14), and enhances invasiveness and migration ability of cells. We found a close association between C3 and TWIST1 in mouse tumors and in normal mouse embryos. Similar to TWIST1, C3 acts as a negative regulator of E-cadherin expression, and might participate in regulating EMT downstream to TWIST1. The effect of C3 on E-cadherin expression is mediated thorough binding of C3a (C3 cleavage product) to its receptor on cancer cells.
Materials and Methods
All of the studies were conducted according to the protocols approved by the Institutional Review Board and Institutional Animal Care and Use Committee of the University of Texas M.D. Anderson Cancer Center.
Cell lines culture conditions
Ovarian and endometrial cancer cell lines (HeyA8, A2780, OVCAR5, SKOV3, ID8-VEGF, Ishikawa, and KLE) were obtained from the institutional Cell Line Core laboratory, where per institutional policy (MD Anderson policy ACA#1044) cell lines are authenticated at least once per year by the short tandem repeat analysis using the Promega Power Plex 16HS kit (Promega) and somatic mutations were detected using a Sequenom MALDI TOF MassArray system (Sequenom). Authentication of the cell lines used in this manuscript was performed within 6 months period prior to the described experiments. Cell lines were routinely genotyped in our lab to confirm identity, and tested by MycoAlert Kit (Lonza) to confirm the absence of mycoplasma. Cell line cultures were incubated in RPMI-1640 or DMEM media with 10-15% FBS and maintained at 37°C in a humidified incubator infused with 20% O2 and 5% CO2.
Quantitative real time PCR (qRT-PCR)
Total RNA was prepared from cells using the Direct-zol™ RNA Miniprep Kit (Zymoresearch). cDNA was synthesized from 1 μg of total RNA using the Verso cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s protocol followed by q-RT PCR using the Power SYBR Green PCR Master mix (Thermo Scientific). Primers were used to detect C3 (Forward; 5′-GCT GAA GCA CCT CAT TGT GA-3′, Reverse; 5′-CTG GGT GTA CCC CCT TCT TGA-3′), TWIST1 (Forward; 5′-TCC ATT TTC TCC TTC TCT GGA A-3′, Reverse; 5′-GTC CGC GTC CCA CTA GC-3′), E-cadherin (Forward; 5′-CCA TCT CAA GCT CGC GGA TA-3′, Reverse; 5′-TCC AAC GTG GTC ACC TGG T-3′ and 18S ribosomal RNA (8).
Chromatin immunoprecipitation (ChIP)
Mouse embryo fibroblasts (MEFs) were grown to near 80% of confluence in 15 cm dishes. Cells were cross-linked with 1% formaldehyde for 10 min at 37°C in RPMI1650. The chromatin was prepared as described previously (15), diluted 10-folds in ChIP dilution buffer, and incubated overnight at 4°C with 2 μg TWIST1 antibody (Twist2C1a) (Abcam) or 2 μg control non-immune IgG (IgG). Immunoprecipitated DNA was purified and used as template in PCR using the primers flanking the identified TWIST1 consensus binding sites: C3 ChIP-TWIST1 Forward-1; 5′ -TCC TCT CCC TCT GTC CCT CT -3′, C3 ChIP-TWIST1 Reverse-1; 5′- GCT AAC CCC TGA ATC CAC AA- 3′, C3 ChIP-TWIST1 Forward-2; 5′- GGG GGT TCT CCA GAC CTT AG- 3′, C3 ChIP-TWIST1 Reverse-2;5′- AAC ATG TCC ATG GGG TGA GT-3′, C3 Non-specific Forward ChIP; 5′ -GAT GGG AGG AAG ACC ACC TT- 3′, C3 Non-specific Reverse ChIP; 5′ -CCC CTC ACT TAC CCT TGT CA- 3′, and 60°C as the annealing temperature to generate a 160-bp amplicon.
Generating C3 promoter-luciferase reporter gene construct
One thousand two hundred and fifty six nucleotides from the C3 promoter (−1005 to +251) was amplified from human blood genomic DNA (Clontech), using Forward primer containing KpnI restriction enzyme cleavage site (5′- GGG GTA CCG AAT ATG CTG TCA ACA GGG ATG -3′) and reverse primer containing BglII restriction enzyme cleavage site (5′- GGA AGA TCT AAC CAC AAA CAC CCA AAC TCA C -3′), and cloned into the pGL3 basic luciferase expression vector (Promega). The QuickChange® IIXL Site-Directed mutagenesis Kit (Stratagene) was used to generate mutant C3 using the C3 luciferase construct as a template and the following primers: forward primer (5′- GCC AGA TAA AAA GCC AGC TCT TTT AGG CGC TGC TCA CTC CTC CC-3’) and reverse primer (5′- GGG AGG AGT GAG CAG CGC CTA AAA GAG CTG GCT TTT TAT CTG GC -3′).
Dual-luciferase reporter gene assay
Luciferase reporter assays were performed as described previously (16). Briefly, HeyA8 cells were plated on 6-well plates (3.5 × 105 cells per well). Twelve hours after plating, the cells were transiently transfected using Fugene HD (Roche) with 1 μg of pC3wt or pC3mut, 0.5 μg of Renilla luciferase plasmid (transfection control), and 0.5 μg plasmids encoding the TWIST1 or 0.5 μg of empty vector (pcDNA3). After 24 hr, cells were harvested and luciferase activity was measured using the Dual-Luciferase Reporter Assay system (Promega) and a Veritas microplate luminometer (Turner BioSystems). Renilla expression was used for normalizing the results.
Incubation of ovarian cancer cells with C3a receptor modulators
Five hundred thousand SKOV3 ip1 human ovarian cancer cells were incubated in serum free media for 2 days with a C3a-receptor agonist peptide (0.2μM) (17) in the presence or absence of a small molecule inhibitor of KLF5 (10μM)(CID5951923, Tocris); or incubated with a C3a-receptor antagonist (0.2μM) (SB290157, EMD Millipore) under the same conditions. A scrambled peptide served as the control. At the end of incubation periods, total mRNA was isolated and processed for E-cadherin qRT-PCR.
Small interfering RNA (siRNA) transfection
Predesigned TWIST1-specific siRNAs were purchased from Sigma Aldrich. Two μg of TWIST1 siRNA was incubated in serum free media (SFM) with 3 μl of lipofectamine (Invitrogen) for 30 min, and the mixture was added into 5 × 105 cells in a 6 well-plate in SFM and incubated for 6 hours. Transfected cells were supplemented with complete media for 2 days.
Lentivirus-mediated delivery of cDNAs and sh RNAs
Murine C3 shRNA, human TWIST1 cDNA, or human C3 cDNA carrying lentiviri were produced by the core facility for Molecular cloning and Lentivirus production system in the Department of Cancer Biology at UT MDACC. Briefly, murine C3 shRNA (or scrambled sequence), human TWIST1 cDNA, or human C3 cDNA was cloned into pGreen Puro-Lentiviral vectors (pGreenPuro™, System Biosciences, Mountain View, CA) and tagged with green fluorescent protein (GFP). Twenty micrograms of these cloned lentiviral vectors were transfected into HEK293T cells along with 15 μg of packaging plasmid (2nd generation psPAX2, Addgene, Cambridge MA) and 15 μg of envelope plasmid (2nd generation pMD2G) using FuGENE transfection reagent (Promega, Madison, WI) in accordance with the manufacturer’s protocol. Supernatants containing the lentivirus were collected, filtered, and added to the cancer cells in the presence of 6 μg/ml Polybrene (Promega). Twenty-four hours later, 4 μg/ml of puromycin was added to the media for 7 days. Transduction efficiency of the cells was calculated by dividing the number of GFP-expressing cells by the total number of cells, and was found to be 100% in all of the experiments. Transduced cells were analyzed by qRT-PCR assay to determine the level of C3 or TWIST1 mRNAs.
Migration and invasion assays
Cells stably overexpressing C3 were detached and plated into 0.1% gelatin-coated or defined matrix (type II human collagen, 0.5 ng/ml laminin and 5 ng/ml gelatin)- coated inserts with 0.8 mm pores for migration or invasion assay, respectively. Cells were incubated for 6 hr for migration assay and 24 hr for invasion assay, fixed, and stained with hematoxyline. Stained inserts were dried for 24 hours and mounted on the slides using xylene-based Permount. Cells were counted in five images taken from each insert at 100 × magnification. Each experiment was conducted in triplicates.
Murine model of ovarian cancer
The syngeneic model of ovarian cancer was generated by intraperitoneal injection of cancer cells. One million murine ovarian cancer cells were resuspended in 200 μl of HBSS and injected into peritoneum of C57BL/6 mice. Eight weeks after injection, mice became moribund and sacrificed. Tumor nodules were resected from peritoneum, counted, and weighed. Some tumor nodules were fixed in formalin and some others saved as fresh frozen samples by embedding in OCT compound.
Western-blot analysis
Western blotting was performed to detect expression of C3, TWIST1 and E-cadherin in ovarian cancer cells. Briefly, cells were lysed with RIPA buffer in the presence of 1 × Roche complete mini protease inhibitor cocktail (Basel, Switzerland), incubated on ice for 30 minutes and centrifuged at 14,000 rpm for 30 minutes at 4°C. Twenty five μl of protein lysis was electrophoresed on 10% SDS–polyacrylamide gel, transferred to nitrocellulose membrane, and incubated with antibodies against C3 (Comp Tech, 1:2000), TWIST1 (Santa Cruz, 1:500) and E-cadherin (BD, 1:500 dilution) overnight at 4°C. Blots were developed using appropriate secondary antibodies and an enhanced chemiluminescence detection kit (ECL, Pierce Biotechnology, Rockford, Illinois). Actin was used as a loading control, and all experiments were performed in duplicate.
Immunostaining
In vitro immunostaining of cancer cells
Ovarian cancer cells were seeded at a density of 4 × 104 cells / well in chamber slides. After 2 days, cells were fixed by 4% paraformaldehyde; blocked and permeabilized by incubation in 1% bovine serum albumin and 0.1% Triton X-100, respectively, for 30 min at room temperature. Subsequently, cells were incubated with anti-C3 (1:200) (Abgent) or anti-E-cadherin (1:200) (Abcam) antibodies overnight at 4°C in a humidified chamber. C3 or E-cadherin was visualized using an anti-rabbit or anti-mouse secondary antibody conjugated to Alexa 594 (1:1000) (Life technologies) followed by counterstaining with ProLong Gold with DAPI (Life technologies). Images were taken at 200 × magnification using Leica DM 4000 B LED microscope (Leica microsystems).
Immunostaining of transverse sections of mouse embryo and tumor
Wild type mouse embryos were collected at ages of E9.5. Implanted tumors were collected 8 weeks after intraperitoneal injection of ID8-VEGF cell stably transduced with C3 shRNA into peritoneum of mice. Immunostaining of C3, TWIST1, and E-cadherin were performed on 4 μm-thick formalin-fixed paraffin-embedded embryos and tumors. Slides were deparaffinized with xylene and decreasing concentrations of ethanol, and rehydrated with PBS. Antigen retrieval was performed using 1 × Borg-decloaker solution (BioCare Medical) or 1 × DIVA solution (BioCare Medical) in a steam cooker at 65° C for 45 minutes, followed by 80 minute cool-down at room temperature. Endogenous peroxidases were blocked with 3% hydrogen peroxide in PBS followed by PBS washes. Non-specific binding was blocked with 5% normal horse serum and 1% normal goat serum in PBS for 20 minutes. Primary antibodies (C3, TWIST1 and E-cadherin) were diluted to 1:200 concentration using 5% normal horse serum (100-200 μL/slide) and incubated overnight at 4° C. After washing slides, biotinylated secondary antibodies were incubated for 20 minutes and amplified using the streptavidin HRP label (4plus Mouse-on-Mouse Avidin-Biotin Detection, Biocare Medical, Concord, CA). In some experiments Alexa-488 anti-rabbit or Alexa-594 anti-mouse secondary antibodies were incubated for 1 hour. After washing with PBS, for immunohistochemistry, the slides were incubated with 100-200 μL of DAB at room temperature, counterstained with hematoxylin for 15 seconds, and mounted on a bright field microscope. For immunofluorescent, DAPI conjugated ProLong® Gold antifade reagent (molecular probes), and mounted on a fluorescent microscope.
Immunostaining of whole-mounted mouse embryos
Wild type mouse embryos were collected at age E11.5. Whole mount staining protocol was provided by Abcam. Briefly, embryos were fixed in 4% paraformaldehyde overnight at 4 degree and washed 3 times in PBS- 1% Triton X-100 (each for 30 minutes). Embryos were incubated in blocking buffer (PBS-1% Triton X-100 + 10% FBS + 0.2% Sodium Azide) at room temperature for a 2-hr period. Endogenous peroxidase activity was blocked by incubation with 0.1% H2O2 diluted in blocking buffer at 4°C overnight. After washes in blocking buffer, embryos were incubated with C3 antibody or TWIST1 antibody for 24 hr at 4°C on a gentle rotation rocker. Embryos were washed 3 times in blocking buffer, each for 1 hr. Sodium azide residue were removed by 3 washes with PBS 1% Triton X-100, each for 10 min. Secondary anti-mouse antibody was diluted in blocking buffer (1:1000, without sodium azide) and incubated with embryos for 2 days at 4°C. After 3 washes in PBS 1% Triton X-100, to detect signal, embryos were incubated in DAB substrate for 2 hours at room temperature, rinsed 3 times in PBS, and incubated in graduate concentration of glycerol (100%, 75% and 50%) before being mounted under a microscope (Leica M80).
Results
C3 promoter has a TWIST1 binding site
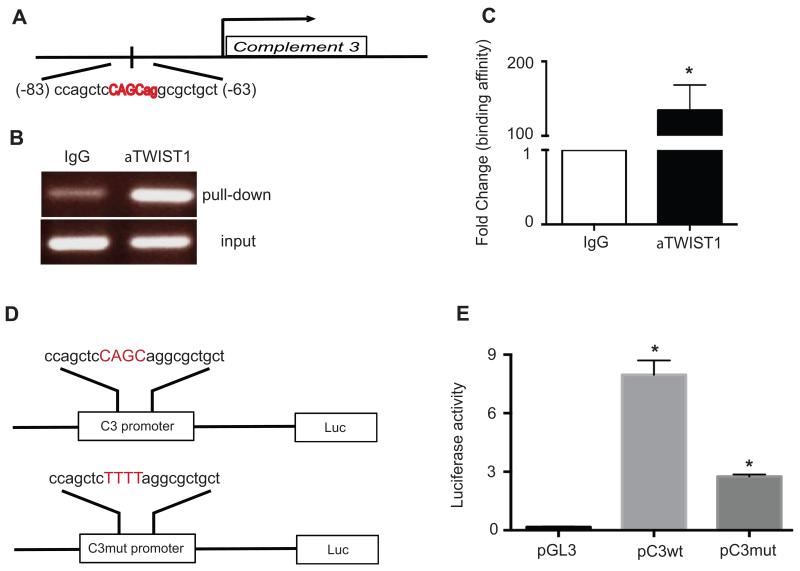
We used promoter-predicting database (Genomatix) and detected two putative TWIST1 binding sites on the C3 promoter (nucleotides 17 → 37 and −83 → −63). To determine whether C3 binds to these sites, we performed chromatin immunoprecipitation (ChIP) using TWIST1 antibody. After ChIP, chromatin DNA was collected and real-time PCR was performed using primers spanning the putative TWIST1 binding sites on the C3 promoter. One of the two sites on the C3 promoter, nucleotide −83 → −63, showed 134-fold higher concentration in chromatin DNA precipitated with anti-TWIST1 antibody than that with control IgG (Fig. 1, A, B and C).
FIGURE 1. TWIST1 binds to the C3 promoter.
(A) A schematic representation of TWIST1 binding site on the C3 promoter (−63 to −83). (B) PCR-amplification products using primers encompassing TWIST1 binding site on DNA template prepared by ChIP with TWIST1 antibody or control non-immune IgG were detected by agarose gel electrophoresis and ethidium bromide staining. (C) Results of qRT-PCR using primers encompassing TWIST1 binding site on the DNA templates prepared by ChIP with TWIST1 antibody or control IgG (n=3, p= 0.0017). (D) A schematic representation of reporter constructs generated by cloning the luciferase gene 3′ to the C3 promoter with either wild type (C3wt) or mutant (C3mut) TWIST1 core binding site. Four nucleotides in the TWIST1 core binding site were altered in C3mut (CAGC → TTTT). (E) HeyA8 ovarian cancer cells were co-transfected with plasmid containing C3 promoter constructs (pC3wt or pC3mut), plasmid containing TWIST1 cDNA, and plasmid containing Renilla luciferase. Promoterless luciferase plasmid (pGL3) was used instead of plasmids with C3 promoter constructs in the control group. Luciferase activity decreased by 66% in HeyA8 cells co-transfected with pC3mut plasmid as compared to that with pC3wt (n=3, p=0.001)
TWIST1 binding to C3 promoter increases expression of the reporter gene
To determine whether binding of TWIST1 to the C3 promoter activates the C3 gene transcription, we generated a construct with 1256 nucleotides of the C3 promoter, including nucleotide −83 to −63, cloned 5′ to a luciferase reporter gene and transfected it to a human ovarian cancer cell line (HeyA8) with a low native C3 expression. In addition to a construct with the wild-type C3 promoter (pC3wt), we generated a mutant construct (pC3mut) that included mutations in the TWIST1 core-binding site (−76CAGC−73 → −76TTTT−73). Plasmid containing C3 promoter constructs (pC3wt or pC3mut) and plasmid containing TWIST1 cDNA (pTWIST1) were co-transfected into HeyA8 ovarian cancer cells. Empty plasmid (containing the luciferase gene with no promoter) was co-transfected with pTWIST1 as a control. Renilla luciferase reporter was used for normalizing the results. Co-transfection of pC3wt with pTWIST1 resulted in 8.3-fold higher luciferase activity compared to that resulted from co-transfection of empty vector with pTWIST1. Mutation in TWIST1 core binding site in the C3 promoter (pC3mut) reduced luciferase activity by 66% as compared to pC3wt construct (Fig. 1, D and E).
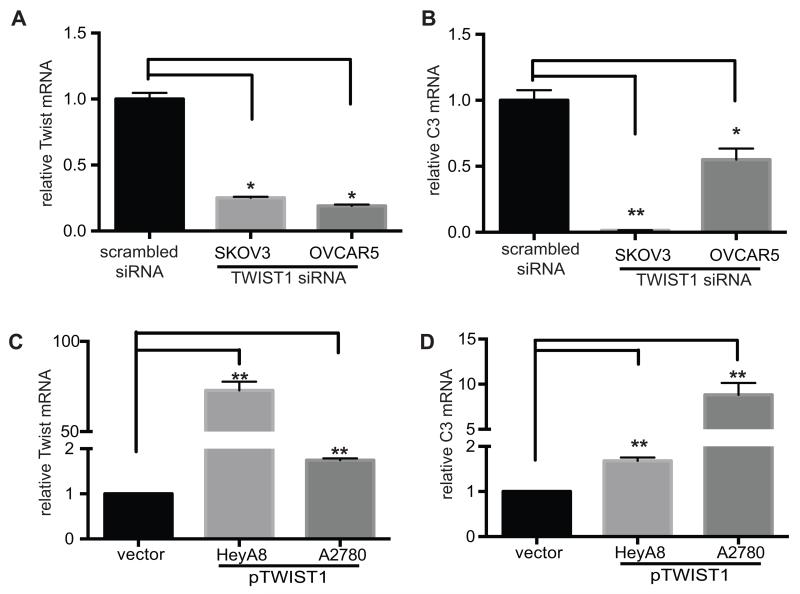
Expression level of C3 parallels expression of TWIST1
Previously we have shown that ovarian cancer cells synthesize C3 (8), and identified different levels of C3 expression in various ovarian cancer cell lines. To study the effect of TWIST1 on C3 expression, we reduced expression of TWIST1 in C3-high ovarian cancer cell lines (SKOV3 and OVCAR5) and increased expression of TWIST1 in C3-low cell lines (HeyA8 and A2780), and measured C3 expression subsequent to TWIST1 manipulation. We reduced expression of TWIST1 using TWIST1 siRNA that resulted in 75% and 78% reduction in TWIST1 expression in SKOV3 ip1 and OVCAR5, respectively. Subsequent to TWIST1 knockdown, C3 mRNA in SKOV3 and OVCAR5 cells decreased by 99% and 48%, respectively (Fig. 2, A and B). We increased expression of TWIST1 in HeyA8 and A2780 ovarian cancer cells by transfecting these cells with plasmid containing TWIST1 cDNA (pTWIST1). Overexpression of TWIST1 increased C3 mRNA in HeyA8 by 1.7 folds and in A2780 by 8.8 folds. (Fig. 2, C and D). These results confirmed that TWIST1 positively regulates the C3 gene expression.
FIGURE 2. TWIST1 expression positively correlates with C3 expression.
(A) Downregulation of TWIST1 by transfecting human ovarian cancer cells (SKOV3ip1 and OVCAR 5) with TWIST1 siRNAs was detected by measuring TWIST1 mRNA. (B) Subsequent to TWIST1 knockdown in C3-high SKOV3ip1 and OVCAR 5 cells, C3 expression was measured by quantitative RT-PCR. TWIST1 knockdown resulted in 99% reduction in C3 expression (n=3, p=0.001). (C) TWIST1 overexpression in C3-low HeyA8 and A2780 ovarian cancer cells by stable transfection of plasmid encoding TWIST1 cDNA (pTWIST1) resulted in 2 folds and 75 folds increase in TWIST1 expression, respectively (n=3, p=0.0001). (D) C3 mRNA levels in HeyA8 and A2780 cells stably transfected with the TWIST1 cDNA was measured by quantitative RT-PCR. TWIST1 overexpression in HeyA8 and A2780 cells resulted in 1.7 and 8.8 folds increase in C3 expression, respectively (n=3, p<0.005)
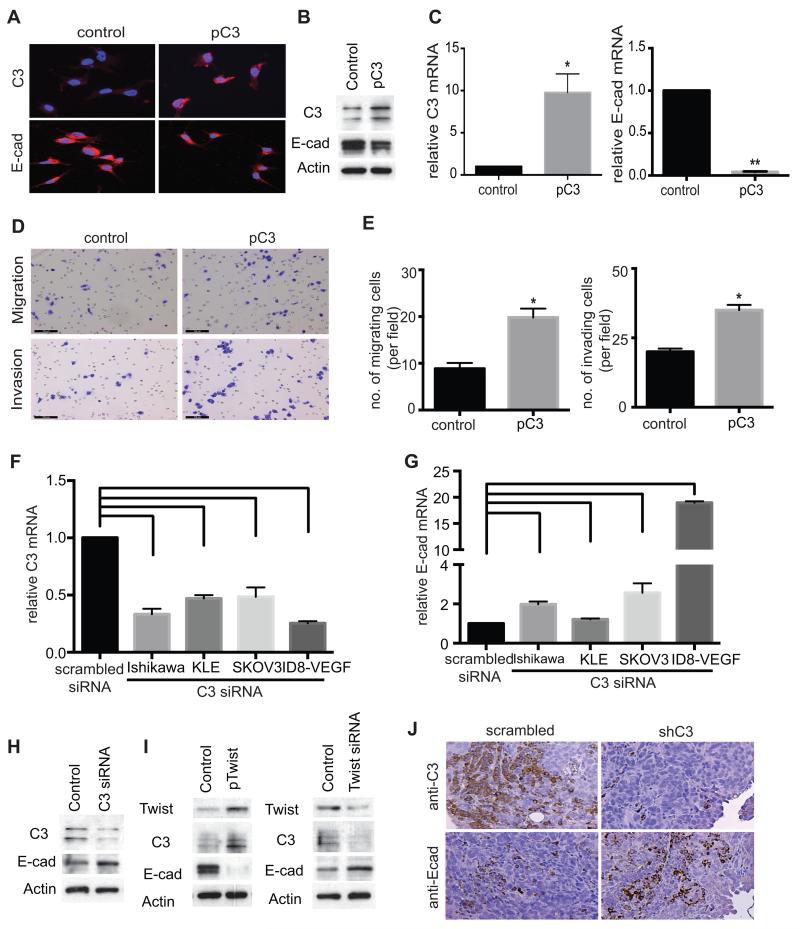
C3 expression negatively regulates E-cadherin expression in vitro and in vivo
TWIST1 is known to promote epithelial-mesenchymal transition (EMT) (18). An important marker of EMT is a reduction in the surface expression of E-cadherin on cancer cells facilitating migration, invasion, and metastasis of these cells. TWIST1 is a transcriptional repressor of E-cadherin, and we investigated whether C3 is also a negative regulator of E-cadherin expression. We overexpressed C3 in HeyA8 ovarian cancer cells by stable transduction of lentivirus carrying the C3 cDNA (pC3). Increase in C3 expression in HeyA8 cells was associated with a reduction in the expression of E-cadherin, as detected by at protein and mRNA level by immunohistochemistry, Western-blotting analysis and qRT-PCR (Fig. 3, A, B and C). To investigate functional consequences of C3 overexpression in vitro, migration and invasion assays were performed using C3-overexpressing HeyA8 cells. C3 overexpression increased migration by 2.3 folds and invasion by 1.8 folds, as compared to control HeyA8 cells transduced with control lentivirus (Fig. 3, D and E). We studied the correlation between C3 and E-cadherin in additional cancer cells lines, using SKOV3 and ID8-VEGF cells (ovarian) and Ishikawa and KLE cells (endometrial). C3 gene knockdown by C3 siRNA increased E-cadherin mRNA level in Ishikawa cells by 2 folds, in KLE cells by 1.3 folds, in SKOV3 cells by 2.5 folds, and in ID8-VEGF by 19 folds (Fig. 3, F and G). Figure 3H shows the results of Western blotting on protein lysate prepared from SKOV3 cell transfected with C3 siRNA or scrambled siRNA and immunoblotting with C3 and E-cadherin antibodies, and is consistent with an increases in E-cadherin after C3 knockdown. To further confirm the effect of manipulating TWIST1 level on expression of C3 and E-cadherin at protein levels in vitro, we overexpressed TWIST1 in HeyA8 cells (low native C3) by transfecting these cells with expression vector carrying TWIST1 cDNA (pTWIST1), and downregulated TWIST1 in SKOV3 cells (high native C3) using TWIST1 siRNA. Western- and immunoblotting showed that an increase in TWIST1 level increased C3 and reduced E-cadherin, and on the other hand TWSIT1 downregulation reduced C3 and increased E-cadherin. (Fig. 3 I).
FIGURE 3. C3 expression negatively correlates with E-cadherin expression in vitro and in vivo.
(A) After stable transduction of lentivirus carrying C3 cDNA (pC3) into low-C3 HeyA8, C3 expression (A, upper panels) or E-cadherin expression (A, lower panels) was detected by immunofluorescence microscopy, (B) by Western-blotting or (C) by quantitative RT-PCR, respectively. HeyA8 cells stably transduced by pC3 showed increase in C3 expression (B, left panel, n=3, p=0.002) and reduction in E-cadherin expression (B, right panel, n=3, p=0.008). (D) The functional effect of C3 overexpression in HeyA8 cells was studied by in vitro migration and invasion assays, using HeyA8 cells stably transfected with pC3. (E) HeyA8 cells overexpressing C3 showed 2.3 folds increase in migration ability (E, left panel, n=15, p=0.001) and 1.8 folds increase in invasion ability (E, right panel, n=15, p=0.001) as compared with those transfected with an empty control plasmid. (F) Ovarian (SKOV3 and OVCAR5) and endometrial (Ishikawa and KLE) cancer cell lines were transfected with C3 siRNA. C3 mRNA level was decreased by 55-75% in ovarian cancer cells and by 52-60% in endometrial cancer cells (n=3, p=0.001) as compared to scrambled siRNA transfected cells. (G) After C3 knockdown E-cadherin mRNA increased by 2.5-19 folds in ovarian cancer cells and by 1.3-2 folds in endometrial cancer cells (n=3, p=0.001). Cell lines transfected with scrambled siRNA served as controls. (H) After transient transfection of C3 siRNA into C3-high SKOV3, C3 and E-cadherin protein expression was determined by Western- and immunoblotting. Scrambled siRNA was used as a control. (I) After stable transfection of HeyA8 cells with plasmid containing TWIST1 cDNA (pTWIST1) and transfection of SKOV3 cells with TWIST1 siRNA, protein expression of TWIST1, C3, and E-cadherin was determined by Western-blotting. (J) Expression of E-cadherin and C3 in sections of ovarian tumors resected from tumor-bearing mice were studied using immunofluorescence microscopy. Tumors induced by ID8-VEGF cells transduced by C3 shRNA showed reduced expression of C3 and increased expression of E-cadherin as compared to tumors induced by ID8-VEGF cells transduced by scrambled shRNA.
We have previously shown that C3 gene knockdown in ovarian cancer cells resulted in smaller tumors in orthotopic murine models of ovarian cancer (8). Wild type C57BL/6 mice injected with ID8-VEGF murine ovarian cancer cells expressing C3 shRNA developed smaller tumors, with 90% reduction in tumor size, as compared to controls (C57BL/6 mice injected with ovarian cancer cells expressing scrambled shRNA). To study the correlation between C3 and E-cadherin expression in vivo, we investigated the effect of C3 gene knockdown on E-cadherin expression in tumors resected from tumor-bearing mice. ID8-VEGF murine ovarian cancer cells expressing C3 shRNA or scrambled shRNA were injected into the peritoneum of C57Bl/6 mice. After 8 weeks, implanted tumors were resected and immunostained for C3 and E-cadherin. While C3 knockdown reduced C3 expression in implanted tumors (Fig. 3 J, upper panels), it was associated with an increase in E-cadherin expression (Fig. 3 J, lower panels).
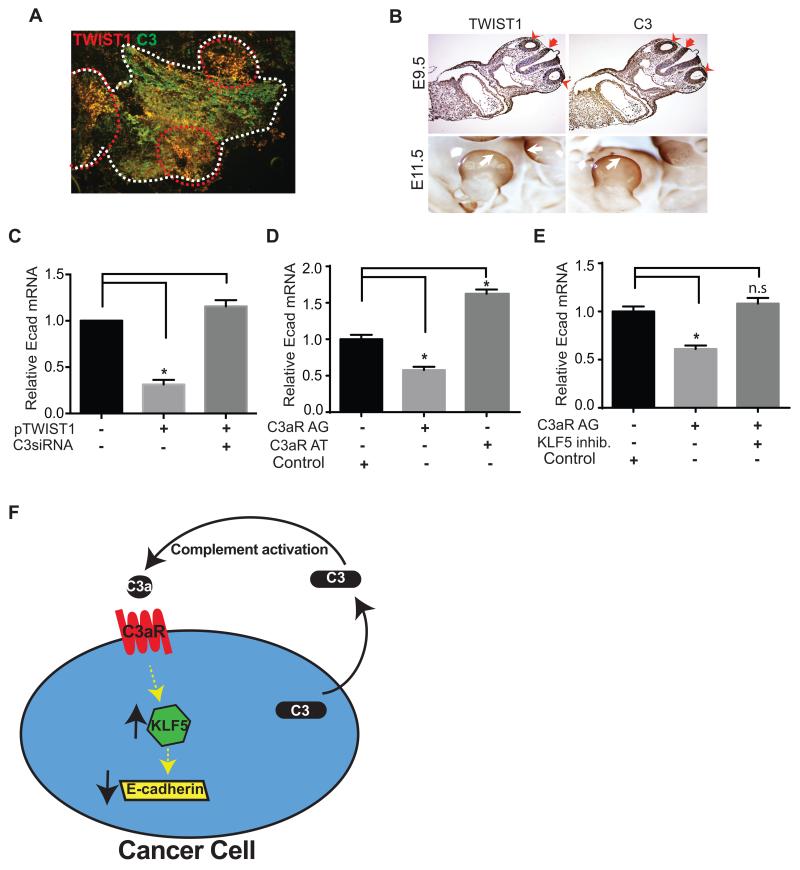
C3 and TWIST1 co-localize in tumor tissue and developing mouse embryos
We investigated C3 and TWIST1 expression at the protein level in tumors by immunofluorescence staining. Tumors induced in mice after intraperitoneal injection of ID8-VEGF murine ovarian cancer cells were resected and immunostained for C3 and TWIST1 proteins. TWIST1 and C3 co-localized at tumor edges, where EMT and tumor cells migration occur (Fig. 4 A).
FIGURE 4. Coexpression of C3 and TWIST1 in implanted tumors in mice and in developing murine embryos.
(A) Expression of TWIST1 and C3 in implanted ovarian tumor tissue was analyzed using immunofluorescence microscopy. ID8-VEGF cells-induced tumors were resected and immunostained with TWIST1 (red) and C3 (green) antibodies. Borders of a tumor nodule are shown using white broken lines. Co-expression of TWIST1 and C3 (yellow) was more prominent at the invasive edges of the tumor (circumscribed with red broken lines). (B) Mouse embryos were collected at 9.5dpc (upper panels) and 11.5dpc (whole mounted, lower panels), and immunohistochemistry was performed using anti-TWIST1 or anti-C3 antibody. TWIST1 and C3 were co-localized in the otocysts (red arrow head) and hindbrain (red arrow) at 9.5dpc embryo, and in the limb-bud (white arrow) at 11.5dpc embryos. (C) To study the effect of C3 on the TWIST1-mediated gene regulation, E-cadherin mRNA level was measured before and after transient transfection of C3 siRNA into TWIST1-overexpressing HeyA8 cells (stably transfected with plasmid encoding TWIST1 cDNA). Scrambled siRNAs were used as negative controls. TWIST1 overexpression was associated with 70% decrease in E-cadherin mRNA level (n=3, *p=0.0002). C3 knockdown restored E-cadherin expression in TWIST1 overexpressing cells. (D) Incubation of SKOV ip1 human ovarian cancer cells with a C3a-receptor agonist (C3aR AG) for 2 days decreased E-cadherin mRNA level by 43% ± 0.1, and incubation with a C3a-receptor antagonist (C3aR AT) increased E-cadherin expression by 62% ± 0.2 (n=3, p<0.0001). Scrambled peptide served as the control. (E) Inhibition of KLF5 abrogated the C3a-receptor agonist-induced E-cadherin suppression (n=3, p<0.0001 and n.s= not significant). Scrambled peptide served as the control. (F) The schematic representation of the role of C3 in a pro-tumor autocrine loop in ovarian cancer. Cleavage of C3 secreted by ovarian cancer cells into tumor microenvironment generates C3a that binds to C3a-receptor on cancer cells. Activation of C3a-receptor increases KLF5, and KLF5 reduces E-cadherin expression.
Given the role of EMT in embryogenesis, we investigate physiologic co-expression of C3 and TWIST1, by immunostaining mouse embryos using antibodies against TWIST1 or C3. Transverse section of 9.5-day post-coitum (9.5dpc) mouse embryos showed co-expression of TWIST1 and C3 in otocyst (ot) and hindbrain (hb). In whole-mounted 11.5dpc mouse embryos, C3 and TWIST1 were co-expressed in developing limb buds (Fig. 4 B).
E-cadherin downregulation by TWIST1 is partly C3-mediated
To investigate whether TWIST1-induced reduction in E-cadherin is C3-mediated or not, we studied the effect of TWIST1 overexpression simultaneous with C3 knockdown, using C3 siRNA in HeyA8 cells stably transduced with TWIST1 cDNA. Overexpression of TWIST1 resulted in 70% reduction in E-cadherin expression in HeyA8 cells that was completely reversed after C3 knockdown (Fig. 4 C).
C3-induced reduction in E-cadherin expression is mediated by C3a and is KLF5-dependent
To identify the mechanism of C3-induced suppression of E-cadherin, we investigated the effect of C3a on ovarian cancer cells. We found that incubation of ovarian cancer cells with a C3a-mimetic peptide (C3a-receptor agonist peptide) (17) decreased expression of E-cadherin in cancer cells, and incubation with a C3a-receptor antagonist increased E-cadherin (Fig. 4D). To investigate the molecular events after C3a-receptor activation, we examined the role of Krüppel-like factor 5 (KLF5) in the C3a-mediated E-cadherin downregulation. KLF5 is a member of zinc-containing transcription factors (19). Activation of C3a-receptor increased KLF5 expression (20,21), and KLF5 downregulated E-cadherin (22). We found that inhibition of KLF5 by a small molecule inhibitor prevented the C3a-receptor agonist-induced reduction in E-cadherin in ovarian cancer cells (Fig 4E). Our data indicate that C3-induced E-cadherin suppression in ovarian cancer cells was mediated by C3a and was KLF5-dependent.
Discussion
C3 is the central component of the complement system, and its expression in hepatocytes is regulated by C/EBPδ and Farnesoid X receptor (FXR) transcription factors (9,23). Although liver is the main source of plasma C3, but C3 is also present in the extravascular space and can be originated from epithelial cells. C3 in the extravascular space participates in several physiologic functions, other than innate immunity, including embryogenesis, organogenesis, and regulating the adaptive immunity; and in several pathologic responses such as in graft-versus-host disease and in enhancing tumor growth. We have recently shown that malignant ovarian epithelial cells secrete C3 that enhances tumor growth in an autocrine fashion (8). The regulation of C3 expression in malignant epithelial cells has not been studied. In the current study, we showed that TWIST1 transcription factor binds to the C3 promoter and enhances C3 transcription.
TWIST1 induces EMT in cancer cells as manifested by downregulation of E-cadherin on cell surfaces. We investigated whether C3 also participate in EMT. Overexpression of C3 reduced E-cadherin, and C3 knockdown increased E-cadherin expression, suggesting that C3 enhances EMT, as was also reflected in the increased invasiveness and migration ability of C3 overexpressing ovarian cancer cells. We studied the expression of C3 and TWIST1 in implanted malignant tumors in mice and detected co-localization of C3 and TWIST1 at the invasive edges of tumor, where EMT is the most active (24).
EMT also occurs in normal tissues, including in migrating neural crest cells during embryogenesis. C3 expression pattern in neural crest of zebra fish embryo mirrors that of TWIST1, and C3 degradation product C3a regulates collective cell migration during embryogenesis (3). We investigated the expression of C3 in murine embryos and detected co-expression of TWIST1 and C3 mouse embryos in hindbrain and otocyst of 9.5dpc embryos and in the limb buds of 11.5dpc embryos. A previous report showed a role for C3 in the mesenchymal transformation of injured renal tubular epithelial cells (25) and EMT of nephrotubulus in mice (21), but our studies is the first report on the presence of a more general role for C3 in EMT in cancer and in normal embryogenesis.
TWIST1 increases C3 expression and reduces E-cadherin expression. We found that C3 also downregulates the E-cadherin expression. We investigated whether TWIST1 effect on E-cadherin expression is mediated through C3. To study this possibility, we overexpressed TWIST1 simultaneously with C3 knockdown in ovarian cancer cells, and observed that TWIST1-induced reduction in E-cadherin was reversible by C3 knockdown. From these results one might conclude that TWIST1 regulatory effect on E-cadherin is partially C3-mediated. We have previously shown that ovarian cancer cells secrete C3 into tumor microenvironment that results in activation of the complement system (8). C3a or anaphylatoxin that is a complement activation product binds to C3a receptor on ovarian cancer cells to complete an autocrine loop. In the current study, we found that activation of C3a receptor decreased E-cadherin expression via a KLF5-dependent mechanism (Fig. 4F). C3-mediated E-cadherin suppression and EMT promotion result in a more aggressive tumor behavior (8). The clinical implication of complement inhibition as a possible therapeutic approach in ovarian cancer requires additional studies.
Acknowledgments
We would like to thank Dr. Elsa R. Flores (Department of Molecular Cellular Oncology, The University of Texas MD Anderson Cancer Center) for providing 9.5dpc embryo paraffin blocks, and Cancer Biology Lentivirus Core Facility of MD Anderson Cancer Center for producing lentivirus.
This work was supported in part by R01CA177909 (to V.A-K. and A.K.S.) and Ovarian Cancer Research Fund (to V.A-K. and A.K.S.).
Footnotes
Disclosure
Authors do not have any potential conflicts of interest.
Reference List
- 1.Mastellos D, Lambris JD. Complement: more than a ‘guard’ against invading pathogens? Trends Immunol. 2002;23:485–491. doi: 10.1016/s1471-4906(02)02287-1. [DOI] [PubMed] [Google Scholar]
- 2.McLin VA, Hu CH, Shah R, Jamrich M. Expression of complement components coincides with early patterning and organogenesis in Xenopus laevis. Int. J. Dev. Biol. 2008;52:1123–1133. doi: 10.1387/ijdb.072465v. [DOI] [PubMed] [Google Scholar]
- 3.Carmona-Fontaine C, Theveneau E, Tzekou A, Tada M, Woods M, Page KM, Parsons M, Lambris JD, Mayor R. Complement fragment C3a controls mutual cell attraction during collective cell migration. Dev. Cell. 2011;21:1026–1037. doi: 10.1016/j.devcel.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strainic MG, Liu J, Huang D, An F, Lalli PN, Muqim N, Shapiro VS, Dubyak GR, Heeger PS, Medof ME. Locally produced complement fragments C5a and C3a provide both costimulatory and survival signals to naive CD4+ T cells. Immunity. 2008;28:425–435. doi: 10.1016/j.immuni.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raedler H, Yang M, Lalli PN, Medof ME, Heeger PS. Primed CD8(+) T-cell responses to allogeneic endothelial cells are controlled by local complement activation. Am. J. Transplant. 2009;9:1784–1795. doi: 10.1111/j.1600-6143.2009.02723.x. [DOI] [PubMed] [Google Scholar]
- 6.Peng Q, Li K, Anderson K, Farrar CA, Lu B, Smith RA, Sacks SH, Zhou W. Local production and activation of complement up-regulates the allostimulatory function of dendritic cells through C3a-C3aR interaction. Blood. 2008;111:2452–2461. doi: 10.1182/blood-2007-06-095018. [DOI] [PubMed] [Google Scholar]
- 7.Pratt JR, Basheer SA, Sacks SH. Local synthesis of complement component C3 regulates acute renal transplant rejection. Nat. Med. 2002;8:582–587. doi: 10.1038/nm0602-582. [DOI] [PubMed] [Google Scholar]
- 8.Cho MS, Vasquez HG, Rupaimoole R, Pradeep S, Wu S, Zand B, Han HD, Rodriguez-Aguayo C, Bottsford-Miller J, Huang J, Miyake T, Choi HJ, Dalton HJ, Ivan C, Baggerly K, Lopez-Berestein G, Sood AK, Afshar-Kharghan V. Autocrine effects of tumor-derived complement. Cell Rep. 2014;6:1085–1095. doi: 10.1016/j.celrep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juan TS, Wilson DR, Wilde MD, Darlington GJ. Participation of the transcription factor C/EBP delta in the acute-phase regulation of the human gene for complement component C3. Proc. Natl. Acad. Sci. U. S. A. 1993;90:2584–2588. doi: 10.1073/pnas.90.7.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruder C, Hagleitner M, Darlington G, Mohsenipour I, Wurzner R, Hollmuller I, Stoiber H, Lass-Florl C, Dierich MP, Speth C. HIV-1 induces complement factor C3 synthesis in astrocytes and neurons by modulation of promoter activity. Mol. Immunol. 2004;40:949–961. doi: 10.1016/j.molimm.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 11.Hong MH, Jin CH, Sato T, Ishimi Y, Abe E, Suda T. Transcriptional regulation of the production of the third component of complement (C3) by 1 alpha,25-dihydroxyvitamin D3 in mouse marrow-derived stromal cells (ST2) and primary osteoblastic cells. Endocrinology. 1991;129:2774–2779. doi: 10.1210/endo-129-5-2774. [DOI] [PubMed] [Google Scholar]
- 12.Fuchtbauer EM. Expression of M-twist during postimplantation development of the mouse. Dev. Dyn. 1995;204:316–322. doi: 10.1002/aja.1002040309. [DOI] [PubMed] [Google Scholar]
- 13.Barnes RM, Firulli AB. A twist of insight - the role of Twist-family bHLH factors in development. Int. J. Dev. Biol. 2009;53:909–924. doi: 10.1387/ijdb.082747rb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Su X, Cho MS, Gi YJ, Ayanga BA, Sherr CJ, Flores ER. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009;28:1904–1915. doi: 10.1038/emboj.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho MS, Chan IL, Flores ER. DeltaNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle. 2010;9:2434–2441. doi: 10.4161/cc.9.12.12051. [DOI] [PubMed] [Google Scholar]
- 17.Langer HF, Chung KJ, Orlova VV, Choi EY, Kaul S, Kruhlak MJ, Alatsatianos M, DeAngelis RA, Roche PA, Magotti P, Li X, Economopoulou M, Rafail S, Lambris JD, Chavakis T. Complement-mediated inhibition of neovascularization reveals a point of convergence between innate immunity and angiogenesis. Blood. 2010;116:4395–4403. doi: 10.1182/blood-2010-01-261503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 19.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao EH, Fukuda N, Ueno T, Tsunemi A, Endo M, Matsumoto K. Complement 3 activates the KLF5 gene in rat vascular smooth muscle cells. Biochem. Biophys. Res Commun. 2008;367:468–473. doi: 10.1016/j.bbrc.2007.12.160. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Fukuda N, Matsuda H, Endo M, Wang X, Saito K, Ueno T, Matsumoto T, Matsumoto K, Soma M, Kobayashi N, Nishiyama A. Complement 3 activates the renal renin-angiotensin system by induction of epithelial-to-mesenchymal transition of the nephrotubulus in mice. Am. J. Physiol Renal Physiol. 2013;305:F957–F967. doi: 10.1152/ajprenal.00344.2013. [DOI] [PubMed] [Google Scholar]
- 22.Shibata M, Chiba T, Matsuoka T, Mihara N, Kawashiri S, Imai K. Kruppel-like factors 4 and 5 expression and their involvement in differentiation of oral carcinomas. Int. J. Clin. Exp. Pathol. 2015;8:3701–3709. [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Pircher PC, Schulman IG, Westin SK. Regulation of complement C3 expression by the bile acid receptor FXR. J. Biol. Chem. 2005;280:7427–7434. doi: 10.1074/jbc.M411473200. [DOI] [PubMed] [Google Scholar]
- 24.Tran DD, Corsa CA, Biswas H, Aft RL, Longmore GD. Temporal and spatial cooperation of Snail1 and Twist1 during epithelial-mesenchymal transition predicts for human breast cancer recurrence. Mol. Cancer Res. 2011;9:1644–1657. doi: 10.1158/1541-7786.MCR-11-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan J, Zhou X, Cui J, Zou Z, Xu Y, You D. Role of complement 3 in TNF-alpha-induced mesenchymal transition of renal tubular epithelial cells in vitro. Mol. Biotechnol. 2013;54:92–100. doi: 10.1007/s12033-012-9547-2. [DOI] [PubMed] [Google Scholar]