Abstract
Our aim is to develop quantitative single-molecule assays to study when and where molecules are interacting inside living cells and where enzymes are active. To this end we present a three-camera imaging microscope for fast tracking of multiple interacting molecules simultaneously, with high spatiotemporal resolution. The system was designed around an ASI RAMM frame using three separate tube lenses and custom multi-band dichroics to allow for enhanced detection efficiency. The frame times of the three Andor iXon Ultra EMCCD cameras are hardware synchronized to the laser excitation pulses of the three excitation lasers, such that the fluorophores are effectively immobilized during frame acquisitions and do not yield detections that are motion-blurred. Stroboscopic illumination allows robust detection from even rapidly moving molecules while minimizing bleaching, and since snapshots can be spaced out with varying time intervals, stroboscopic illumination enables a direct comparison to be made between fast and slow molecules under identical light dosage. We have developed algorithms that accurately track and co-localize multiple interacting biomolecules. The three-color microscope combined with our co-movement algorithms have made it possible for instance to simultaneously image and track how the chromosome environment affects diffusion kinetics or determine how mRNAs diffuse during translation. Such multiplexed single-molecule measurements at a high spatiotemporal resolution inside living cells will provide a major tool for testing models relating molecular architecture and biological dynamics.
Keywords: Living cells, single particle tracking, super-resolution imaging, co-movement analysis, intracellular labeling
INTRODUCTION
Most cellular processes are the result of multiple factors coming together at precise subcellular locations to form complexes that turnover extremely fast. Information processing in the cell via replication, transcription and translation is orchestrated by an intricate interplay among substrates (DNA for replication and transcription, mRNA for processing and translation), enzymes (DNA and RNA polymerases and the ribosome) and accessory regulatory factors (transcription and translation factors). In order to understand any of these processes on the quantitative level in vivo it is necessary to follow at least three of the actors involved: the substrate, the enzyme and the regulator, in living cells. In eukaryotes, multiple factors bind to the gene promoter region to initiate transcription: general transcription factors (TFs), numerous sequence specific TFs and co-activators, and RNA Polymerase II (Pol II). Enhancer-regulated expression involves additional interactions with distant DNA sequences. With the exception of elongating Pol II, association of these factors has been shown to last for seconds or less. The lack of methodology capable of imaging several of these factors simultaneously in live cells with a time resolution that captures short-lived intermediates seriously impairs our prospects of gaining deeper insight of transcription initiation, and of developing quantitative models of how transcription initiation is regulated. We have recently demonstrated our ability to follow single molecules of two distinct species (mRNA and the nuclear pore complex) in live cells with high spatiotemporal resolution (20 ms and 20 nm, respectively) [2]. Based on these experiments, we are now able to draw a picture of nuclear export at an unprecedented level of detail. Although two-color single molecule technology is extremely powerful, it is still falling far short of the typical number of factors involved in most biological processes. The main objective was to develop a microscope capable of interrogating three components simultaneously at the single molecule resolution. We present a three-camera imaging microscope with synchronized stroboscopic excitation for fast tracking of multiple interacting molecules at the single molecule level, with high spatiotemporal resolution. Our multi-color super-resolution approach has allowed for automated detection and analysis of specific binding and unbinding events of multiple different interaction partners, thus providing insights into the multi-step kinetics of information processing in the cell.
METHODOLOGY
We designed and constructed a custom-built microscope around the easily accessible and adaptable ASI Rapid Automated Modular Mounts (RAMM) frame (purchased from BioVision Technologies) using three separate tube lenses and custom multi-band dichroics to allow for enhanced detection efficiency. The frame times of the three Andor iXon Ultra EMCCD cameras are hardware synchronized to the laser excitation pulses of the three excitation lasers, which illuminate cells in either objective-type total internal reflection fluorescence (TIRF), highly inclined and laminated optical sheet (HiLo), or wide-field (Epi) configuration. This design around the easily accessible RAMM frame (ASI) requires no relay lenses, shutters, or AOTFs (Acousto-Optic Tunable Filters) for optimal imaging with minimal loss. The microscope has no moving parts, which makes the system inherently stable.
2.1 The excitation path of the 3-color microscope
Custom multi-band dichroics and filters allow for more flexibility and enhanced detection efficiency. The system is built around a custom-built three-camera microscope using an Olympus 1.4 NA PLAPON 60x OSC objective (or an Olympus 1.49 NA 60x TIRF objective for TIRF illumination). The microscope has an upper and a lower port (C60-RA-2nd- PORT, ASI). A general purpose mirror (MGP01-650-1300-25x36, Semrock) is taped but not glued (to avoid strain) into the C60-RA-MIRROR assembly (ASI) and reflects light below 800nm into the top port, but provides a downward path for the 940 nm light emitted by the diode of the ASI CRISP autofocus device. The CRISP system is mounted on the lower port and provides continuous focus stabilization. The C60-RA-MIRROR assembly is attached to the LS-50 focus actuator, which is used by the CRISP system for focus stabilization, as well as for large-scale movement of the objective. For long-term imaging, the CRISP autofocus system based on the reflection of the 940nm diode provides continuous and simultaneous positional feedback along the optical axis. The fluorescence path is placed on the top port of the 3-color microscope to keep the fluorescence infinity space as short as possible as the optical infinity path of the lower port is 75 mm longer than the upper port. Four laser lines (407, 490, 555 and 639 nm) from a Stradus 405-100, a Stradus 488-150, a Stradus 637-140 (all Vortran) and a 555-nm DPSS laser (CL555-1000-O with TTL Modulation, CrystaLaser) are combined using four 90o periscopes for easy alignment. The red 639 nm laser is coupled in using an end mirror, and a ZT561rdc, a ZT532rdc, and a ZT405rdc round 25mm diameter dichroics (all Chroma) are used to couple in the remaining lasers. The combined laser beams are expanded 3x, and passed through a tilted rotating ground glass 1o diffuser (Süss MicroOptics). A TIRF lens (LAO-250.0-25.0, Melles Griot) focuses the laser light onto the back aperture of the objective. The lens is mounted in a 60mm cage system (Thorlabs) and translates together with a broad-band end mirror at right-angle (⌀50.8mm, BB2-E02, Thorlabs) as part of a 90o periscope to achieve seamless transitioning from Epi, HiLo and TIRF illumination using one dial. HiLo allows for both optical sectioning and fast imaging with background fluorescence rejection at a very low cost and is especially useful when imaging in the nucleus since it achieves excellent signal to noise ratio. The laser light is coupled into the microscope by an adjustable cube (MIM- CUBE-II-K, ASI) holding a factory-mounted multi-band dichroic (zet405/488/561/635x-WF, Chroma) to filter the laser light. All dichroics are factory mounted (and interferometry tested for flatness) into TIRF cube holders (Laser TIRF Cube for Olympus BX/IX, Chroma). The MIM-CUBE-II-K allows for both precision alignments as well as quick- interchange between dichroic holders using magnetic locks. This is crucial for the 3-camera system, as precision alignment of the excitation as well as the emission paths is essential. This first MIM-CUBE-II-K cube also holds a multi- band excitation filter (zet405/488/561/635x-WF, Chroma) to filter the four laser lines, and a quad-notch filter (NF01-405/488/557/640-25x5.0-D, Semrock), which further filters the emitted light (see Figure 1).
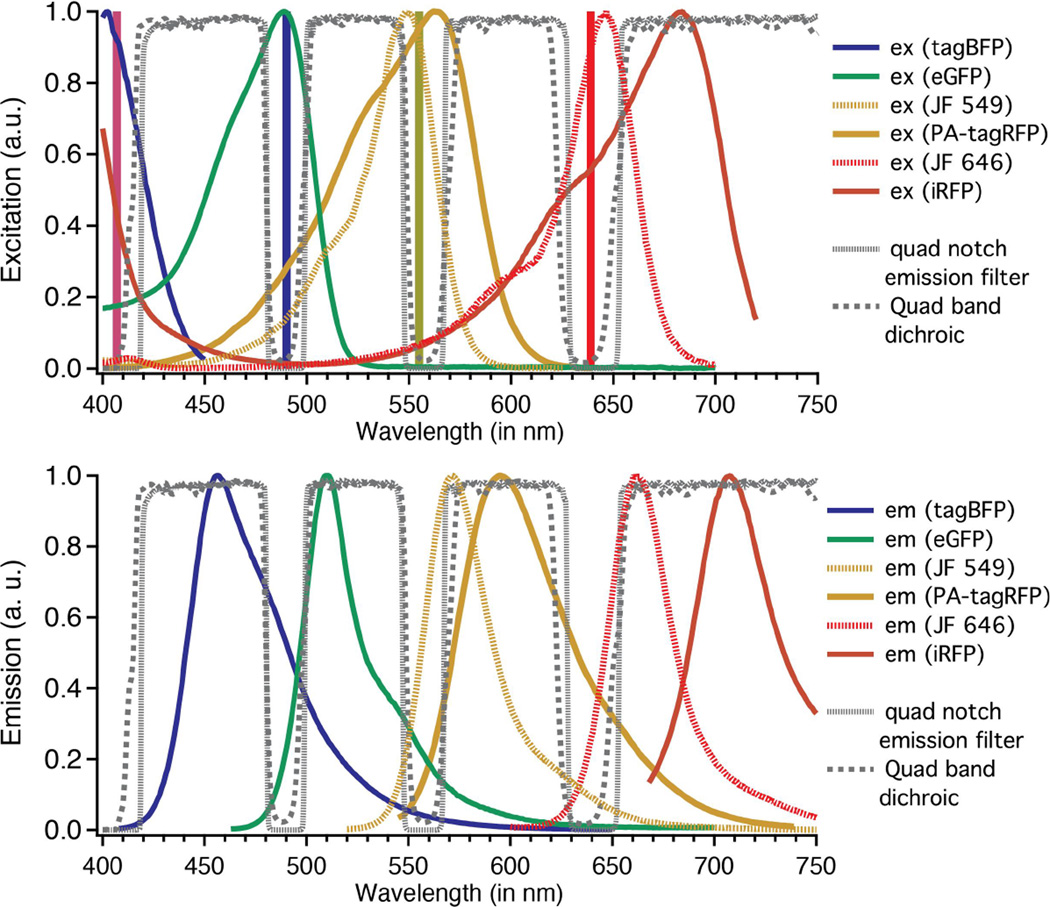
Figure 1.
The four emission windows of the 3-camera microscope with excitation and emission spectra of suitable fluorophores. The vertical lines highlight the four laser lines. A suitable dye combination for simultaneous single- molecule imaging is eGFP, JF 549 and JF 646 [1].
2.2 The emission paths of the 3-color microscope
The multi-band optics allows for detection of four open emission windows (420 – 480; 500 – 550; 565 – 630; and 650 – 800 nm, Figure 1), and three simultaneously without moving parts (see Figure 2), which greatly enhances drift stability of the microscope. A second MIM-CUBE-II-K cube on the top port splits the ‘blue’ emission path: it holds a 2-mm-thick emission dichroic (T490pxrxt-A/R coated, Chroma), and a bandpass emission filter (FF01-525/45-25, Semrock). A third MIM-CUBE-II-K cube on the top port holds a 2 mm thick emission dichroic (T560lpxr – lot 253099, Chroma), and splits the ‘orange’ emission channel, which is filtered further by a band-pass emission filter (FF01-609/54-25, Semrock). A bandpass filter (FF01-731/137-25, Semrock) filters the transmitted emitted light of the third, ‘far red’ emission channel. Three images are formed using the Olympus objective and three custom tube lenses that are placed in the infinity space of each of the three channels (LAO-300.0, Melles Griot), resulting in 100x overall magnification.
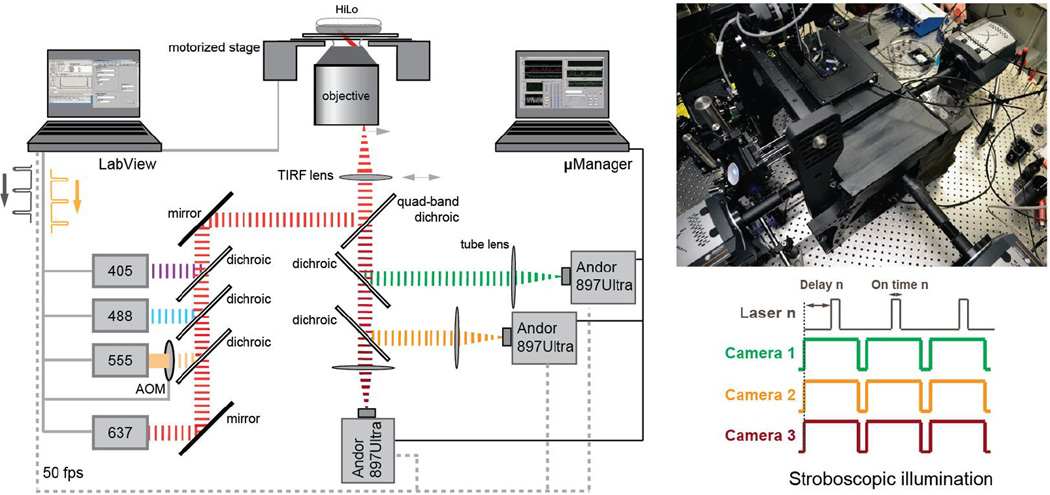
Figure 2.
Diagram and image of the optical path of the 3-color microscope. The cameras are hardware synchronized to obtain simultaneous images on three cameras. All lasers are strobed with independent delays and on-times.
2.3 Laser and camera hardware synchronization and camera control
Three iXon Ultra 897 EMCCD high-speed Andor cameras are synchronized using a National Instruments DAQ board (NI-DAQ-USB-6363) with four independent counters. All four lasers allow for direct TTL modulation, and millisecond stroboscopic excitations are synchronized to the frame times of the respective cameras via LabVIEW 2012 (National Instruments): The 490 nm laser is synchronized with the frame-rate of the first camera (blue emission path), the 555nm laser is synchronized with the second camera (orange path), and the 639 nm laser is synchronized with the third camera (far-red path). All lasers stroboscopically illuminate the sample with independent delay and on-times using peak power levels around 1 kW/cm2 at the sample (see Figure 2). Stroboscopic illumination allows robust detection of distinct fluorescence spots from even rapidly moving objects, and hence accurate single particle tracking is possible even for freely diffusing proteins [3] in all three channels. Additionally, the stroboscopic illumination scheme minimizes photobleaching, and hence allows for the recording of longer single particle trajectories. The fourth counter of the DAQ board uses an on-board 100 KHz time-base to trigger and strobe the 407 nm photoconversion and photoactivation laser: Photoconvertible protein molecules such as Dendra2 can be photoconverted by 100 μs long excitation pulses of 407 nm (50 W/cm2) light every 200 ms during the course of image acquisition. Fluorescence is detected using three liquid- cooled (EXT-440 re-circulator, Koolance) back-illuminated EMCCD Andor Ixon Ultra cameras (cooled to −80 oC, with 17 MHz EM amplifiers). Two of the cameras have sensors with standard AR coating (for the blue and orange channels: model DU-897-CS0-BV), and the third camera has a coating with increased sensitivity in the NIR (model DU-897U- CS0-EXF, for the far-red channel). A counter of a second NI-DAQ-USB-6363 acquisition board is used to synchronize the three cameras (either in External Start or External Exposure trigger modes). All three cameras are controlled simultaneously using μManager (1.4.20) [4]. A Dell Precision T7600 computer with two six-core Xeon processors (E5- 2630, 2.3 GHz, Intel) and 64 GB of DDR3 RDIMM memory (1600 MHz) controls μManager. Two additional USB 3.0 adapter cards (PEXUSB3S2, StarTech) were installed to ensure that each of the three cameras has a unique and unshared USB controller, which enables uninterrupted continuous streaming of all three cameras simultaneously at 50 Hz in full- frame mode or at 100 Hz with a central region of interest with 256x256 pixels. A second computer controls the two DAQ boards in charge of hardware timing and synchronization.
2.4 Emission path alignment
The emission path is mapped using a silver reflective collimator with a Ø8.5 mm beam width (RC08APC-P01, Thorlabs) that is placed in place of the objective using a RMS thread adapter (SM05RMS, Thorlabs). The reflective nature of the collimator minimizes chromatic aberrations so that the three emission paths can be accurately mapped simultaneously. White light (490-700 nm) is coupled in via a SuperK broadband fiber (A502-020-010, NKT Photonics) using a white light laser source (SuperK Extreme EXW-12, S442-125-000, NKT Photonics). We place the three tube lenses (LAO- 300.0, Melles Griot) into the exact center of the emission paths by aligning their back-reflections of the laser light. We align the XY and Z-positions of the three cameras by imaging the focused laser spots directly after blocking most of the light using several neutral density (ND) filters (OD=8). The cameras are free-space mounted using 3-axis stages (562 ULTRAlign, Newport) and high-capacity actuators (HR-13, Newport) and a custom-made base plate (see Figure 3).
Figure 3.
Diagram of the 3-axis camera mount with custom base plate for extra drift stability.
2.5 Stage and environmental control for live-cell imaging
Two translation stages, a XY-coarse translation stage (M-545K003 with C-867.26x controller, Physik Instrumente) and a piezo XYZ-top stage (P-545.3C7 with E-545.3CD PInano Piezo Controller, Physik Instrumente) hold a Tokai-hit stage- top incubator (INUP-PPZI-F1 with a built-in analog gas flowmeter). The Z-piezo allows us to incorporate rapid volumetric z-sweep scanning for rapid imaging of entire nuclei using HiLo illumination: stepping through a sample using four 0.5 μm steps can generate z-stacks through the entire nucleus. HiLo microscopy and rapid volumetric z-sweep scanning of the entire nucleus minimize phototoxicity and photobleaching. During imaging, cells are maintained at 37 °C using the Tokai-hit stage-top incubator and objective heater. Two gas tanks were installed outside the laser room to allow for a 5% CO2 atmosphere in the sample chamber (the first gas tank contains a mixture of 5% CO2, 3% O2, 92% N2 for low-oxygen imaging conditions, and a second tank contains 5% CO2 and 95% Air).
2.6 Image registration
Slide-mounted TetraSpeck Fluorescent Microspheres (T14792, Invitrogen) are imaged before and after data acquisition. Short movies (100 frames) of the fluorescent broadband beads in all three channels are registered using the similarity (2d) transformation model using the descriptor-based series registration (2d/3d+t) Fiji plugin [5]. The three channels have to be set as timepoints for this to work for multiple channels (in Fiji: Image>Hyperstacks>Re-order Hyperstack). The three-channels are co-registered with an average error of less than 10 nm full-chip.
2.7 Particle localization, tracking and analysis
For PALM and STORM imaging, Localizer [6] is used for 8-way adjacency particle detection with 20 GLRT sensitivity and a PSF of 1.3 pixels. The resulting particles are drift corrected using fiducial markers. For each detected particle, integrated fluorescence intensities are converted to photon counts using analysis routines written in Igor Pro version 6.36 (WaveMetrics) [1]. The mean and median localization errors are determined using Equation 6 in reference [7].
Tracking analysis is performed using the commercial tracking software DiaTrack (v. 3.03, Semasopht) [8], which identifies and fits the intensity spots of fluorescent particles with 2D Gaussian functions according to experimentally determined point-spread functions. The trajectories are also analyzed with HMM-Bayesian inference, which infers diffusive and directed motion states from observed particle displacements and annotates when and where each motion state occurs in space and time along each trajectory with single-step resolution [9].
RESULTS
We utilized two-channel combinations of our 3-color microscope to characterize the single-molecule properties of a novel photoswitchable long-stokes shift fluorescent protein developed in the Verkhusha laboratory [10], the multivalent spaghetti-monster probes developed in the Looger lab [11], as well as a whole variety of bright and spectrally distinct set of dyes developed in the Lavis laboratory [1]. The cell-permeable nature of the Janelia Fluorophores (JF) has enabled a whole set of live-cell multi-color tracking experiments that would otherwise not have been possible. Most importantly, this new color palette of bright and photostable dyes is crucial to single-molecule tracking experiments within the nucleus of living cells. This new palette, in combination with our HiLo multi-laser setup that can selectively illuminate the nucleus of a cell, has allowed the study of how DNA-binding enzymes are regulated within the chromosomal environment and how this affects transcriptional output. In an ongoing collaboration with Prof. Ibrahim Cissé (MIT), Dr. Luke Lavis (Janelia) and Dr. Timothée Lionnet (Janelia Transcription Imaging Consortium) we track the transcriptional output of a gene for tens of minutes without bleaching, while simultaneously imaging Pol II clustering dynamics at the gene locus at sub-diffraction limited resolution. We employ the MS2 fluorescent reporter system [12] to label the endogenous β-actin mRNA, and simultaneously capture Pol II cluster dynamics [13] specifically at nascent transcription sites. We use MEF cells derived from a mouse where the bacteriophage MS2 stem loops had been knocked into the endogenous β-actin gene [14] and that express the α-amanitin resistant Dendra2-Pol II construct [13] as well as the halo- tagged MS2 coding protein (halo-MCP). The halo-MCP is labeled with a novel near-infrared, cell-permeable and fluorogenic Janelia Fluor 700 dye developed in the Lavis lab, which allows for PALM imaging of both Dendra2-Pol II as well as β-actin mRNAs without cross-talk and cross-bleaching effects of the two fluorophores [15].
CONCLUSION AND OUTLOOK
Single-color single-molecule studies of biological factors have recently become possible inside the nucleus of living eukaryotic cells [16]. Such experiments have proven extremely valuable in quantitatively testing models of diffusion and target search. However, one-color experiments provide limited access to the complexity of multicomponent interactions such as occur during transcription or translation. We have applied multi-color super-resolution approach to study transcriptional enzymes and regulators in the nuclei of living cultured cells. The tools developed for studying cells in culture are applicable also to translational processes in acute tissue slices, and ultimately in an in vivo context. Furthermore, our multi-color imaging methodology has the potential to go beyond studying modifications at a single locus (e.g., the β-actin locus). It is in principle capable of globally assessing the modification states at many loci at the same time, over timescales of many minutes. We anticipate that this method will be an important tool to uncover the spatiotemporal organization of the genome as a whole in the cell nucleus. While there has been important recent progress in our understanding of chromatin architecture thanks to 3C-related ensemble techniques, our imaging-based single-cell approach provides a powerful complementary assay to directly address dynamics and heterogeneity. This global approach is challenging from an imaging perspective since chromosomes are not static structures over the minute timescales; the chromosomal architecture within a nucleus is a dynamic entity. This is not of concern if the gene locus is marked with a beacon (as is the case with MCP tagging of the β-actin locus [15]), however it will not be possible to develop beacons for all gene loci of interests simultaneously, and the dynamic rearrangements will have to be accounted for if modifications at the genome scale can be tracked for many minutes. With the advent of new genetic engineering techniques based on the CRISPR/Cas system, molecular enzymes as well as DNA [17] can be specifically labeled with the fluorescent tags used to achieve high spatial and temporal resolution in living cells. It is now technically feasible to develop a CRISPR/Cas9 guide-star system where fluorescent puncta will serve as internal reference points. This will allow for the movement of loci to be accounted for on the minute timescale as the chromosome is dynamically rearranging. Multicolor quantitative imaging and analytical approaches will enable a wide range of studies probing nuclear organization and dynamics, with unprecedented spatial and temporal resolution directly in living cells. The three-color single–molecule approach is general in nature; any pair of interacting factors can in principle be labeled and imaged at the single molecule level, while global dynamics can be accounted for simultaneously in the third channel. This will provide a powerful tool to directly assay spatiotemporal dynamics and unravel cause-and-effect relationships across the cell.
Acknowledgments
We thank David Grünwald and Timothée Lionnet for their technical expertise and helpful discussions, and Igor Negrashov for his manufacturing expertise at the Janelia Instrument Design and Fabrication Department. We acknowledge our Pol II clustering collaborator Ibrahim I. Cissé, and we thank Jonathan B. Grimm and Luke D. Lavis for the development and synthesis of Janelia Fluor 700. The Howard Hughes Medical Institute supported this work.
REFERENCES
- 1.Grimm JB, English BP, Chen J, et al. A general method to improve fluorophores for live-cell and single- molecule microscopy. Nat Methods. 2015;12(3):244–250. doi: 10.1038/nmeth.3256. 3 p following 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grunwald D, Singer RH. In vivo imaging of labelled endogenous beta-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467(7315):604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.English BP, Hauryliuk V, Sanamrad A, et al. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(31):E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edelstein A, Amodaj N, Hoover K, et al. Computer control of microscopes using micromanager, Chapter 14, Unit14 20. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preibisch S, Saalfeld S, Schindelin J, et al. Software for bead-based registration of selective plane illumination microscopy data. Nat Methods. 2010;7(6):418–419. doi: 10.1038/nmeth0610-418. [DOI] [PubMed] [Google Scholar]
- 6.Dedecker P, Duwe S, Neely RK, et al. Localizer: fast, accurate, open-source, and modular software package for superresolution microscopy. J Biomed Opt. 2012;17(12):126008. doi: 10.1117/1.JBO.17.12.126008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortensen KI, Churchman LS, Spudich JA, et al. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat Methods. 2010;7(5):377–381. doi: 10.1038/nmeth.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vallotton P, Olivier S. Tri-track: free software for large-scale particle tracking. Microsc Microanal. 2013;19(2):451–460. doi: 10.1017/S1431927612014328. [DOI] [PubMed] [Google Scholar]
- 9.Monnier N, Barry Z, Park HY, et al. Inferring transient particle transport dynamics in live cells. Nat Meth. doi: 10.1038/nmeth.3483. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piatkevich KD, English BP, Malashkevich VN, et al. Photoswitchable red fluorescent protein with a large Stokes shift. Chem Biol. 2014;21(10):1402–1414. doi: 10.1016/j.chembiol.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viswanathan S, Williams ME, Bloss EB, et al. High-performance probes for light and electron microscopy. Nat Methods. 2015;12(6):568–576. doi: 10.1038/nmeth.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertrand E, Chartrand P, Schaefer M, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2(4):437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 13.Cisse II, Izeddin I, Causse SZ, et al. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341(6146):664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 14.Lionnet T, Czaplinski K, Darzacq X, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho WK, Jayanth N, English BP, et al. Mechanism of Pre-Transcriptional Regulation Through The Spatiotemporal Modulation of RNA Polymerase II in Living Cells. submitted. [Google Scholar]
- 16.Chen J, Zhang Z, Li L, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156(6):1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Gilbert LA, Cimini BA, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155(7):1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]