Abstract
Leiomyosarcoma (LMS) of soft tissue is a sarcoma with smooth‐muscle differentiation, and conventional chemotherapy does not improve its outcome. The application of novel antitumor agents and precise prognostication has been demanded. The expression of the protein Forkhead box M1 (FOXM1), a member of the FOX family, is considered an independent predictor of poor survival in many cancers and sarcomas. However, the expression status of FOXM1 in LMS is poorly understood. The purposes of this study were to examine the correlation between the expression of FOXM1 and clinicopathologic or prognostic factors and to clarify the efficacy of FOXM1 target therapy in LMS. We evaluated the immunohistochemical expressions of FOXM1 using 123 LMS tumor specimens. Univariate and multivariate survival analyses revealed that FOXM1 expression was associated with poor prognosis in LMS. An in vitro study was then carried out to examine the antitumor effect of a FOXM1 inhibitor (thiostrepton) and siRNA on a novel LMS cell line, TC616. We also assessed the efficacy of the combined use of doxorubicin and thiostrepton. Thiostrepton showed dose‐dependent antitumor activity and TC616 cells treated with the combination of thiostrepton and doxorubicin showed lower proliferation compared to those treated with either drug individually. FOXM1 interruption by siRNA decreased cell proliferation and increased chemosensitivity. In conclusion, FOXM1 has potential to be a therapeutic target for LMS.
Keywords: FOXM1, immunohistochemistry, leiomyosarcoma, prognosis, thiostrepton
Soft tissue leiomyosarcomas (LMS), which account for 10–15% of soft tissue sarcomas, are defined as malignant tumors showing smooth‐muscle differentiation.1 Leiomyosarcomas are cytogenetically highly complex tumors without recurrent chromosomal aberrations.2 The main current treatment method for LMS is surgical resection, with or without radiotherapy, but the prognoses of metastatic cases and unresectable cases are still poor.3 The application of novel antitumor agents and precise prognostication are essential to improve the survival of individuals with LMS.
The protein Forkhead box M1 (FOXM1), a member of the FOX family of transcription factors, is widely expressed in embryonic tissues.4, 5 Terminally differentiated non‐proliferating tissues display relatively low levels of FOXM1 expression.6 FOXM1 regulates a wide spectrum of tumor progression processes.7 Increased levels of FOXM1 expression have been detected in many different types of human cancers8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 and sarcomas such as rhabdomyosarcoma,19 Ewing sarcoma,20 malignant peripheral nerve sheath tumor,21 and osteosarcoma.22, 23 Silencing FOXM1 expression suppressed the proliferation of both cancer13, 15, 18 and sarcoma cell lines.19, 23
In various carcinoma cell lines, FOXM1 was also shown to be involved in resistance to chemotherapy drugs such as doxorubicin (DOX)24 which is a frequently used antitumor agent against soft tissue sarcoma. The inhibition of FOXM1 thus has the potential to be a therapeutic target for many malignancies. In LMS, the prognostic impact of FOXM1 expression and the effectiveness of FOXM1 inhibition remain to be clarified.
We carried out a clinicopathologic and prognostic analysis of FOXM1 expression in a series of 123 LMS clinical specimens. We then tested the antitumor activity of a FOXM1 inhibitor (thiostrepton) and siRNA on a soft tissue cell line that originated from LMS tissue.
Materials and Methods
Patients and clinical information
We used samples of soft tissue LMS registered in the Department of Anatomic Pathology, Graduate School of Medical Sciences, Kyushu University (Fukuoka, Japan). Each tumor was classified according to its location and histology by reference to the most recent WHO classification.1 The tumor locations were categorized into somatic soft tissue (proximal or distal), retroperitoneum, and large vessels. Leiomyosarcoma samples from the abdominal cavity or external genitals were excluded from this series.
All of the cases were reviewed based on histological examinations with H&E staining and on an immunohistochemically positive reaction of at least two of the following markers: α‐smooth muscle actin, desmin, and muscle‐specific actin. When the LMS patient was treated with chemotherapy before resection, we examined the patient's corresponding biopsy specimens. Histological grade was evaluated according to the grading system of the French Federation of Cancer Centers Sarcoma Group (FNCLCC).1 For the staging of the primary tumors, the latest American Joint Committee on Cancer staging system was used.25
Survival data were available for overall survival (OS) in 108 patients (87.8%) who had a follow‐up ranging from 0 to 346 months (median, 65 months) and a 5‐year OS rate of 55.9%. Data were also available for event‐free survival (EFS) in 107 patients, who had a follow‐up ranging from 0 to 278 months (median, 33 months). We also analyzed the FOXM1 expression and OS rate in 28 patients who had undergone pre‐ and/or post‐operative chemotherapy.
This study was carried out in accordance with the principles embodied in the Declaration of Helsinki, and was approved by the Ethics Committee of Kyushu University (No. 26‐49).
Cell line
The original tumor tissue specimen was surgically obtained from a soft tissue LMS of a 26‐year‐old man that arose in the chest wall, diagnosed as described above. Fresh tumor tissue was minced and seeded in a 25‐cm2 plastic flask containing DMEM with 10% FBS and penicillin and maintained in a humidified atmosphere of 5% CO2 in air at 37°C. When semiconfluent layers were obtained, the cells were dispersed with PBS containing 0.1% trypsin and 0.02% EDTA solution and seeded in new flasks for passage. We named this cell line “TC616.” After 100 passages, we carried out the assays described below.
Immunohistochemical study of clinical samples
Formalin‐fixed paraffin‐embedded samples of soft tissue LMS from 123 patients were prepared for the immunohistochemical study. These samples had been obtained from biopsy specimens or surgically resected tumors. Samples after chemotherapy were not included. All 123 sections were formalin‐fixed, paraffin‐embedded tissue cut at 3‐μm thickness. Antigen retrieval was carried out by boiling slides with target retrieval solution (Dako, Carpinteria, CA, USA). The primary antibody was monoclonal anti‐human FOXM1 antibody (R&D Systems, Minneapolis, MN, USA) diluted at 1:300. All immunocomplexes were visualized by the Dako EnVision System detection system.
For FOXM1, immunoreactivity was defined as cells showing nuclear staining with/without cytoplasmic staining patterns in the tumor tissue with minimal background staining. Coexistent endothelial cells were evaluated as a negative internal control. Immunoreactivity for FOXM1 was defined based on a previously reported method as follows. Tumors with a strong staining intensity in >10% of the tumor cells were recorded as being positive.8, 9 The serial sections were also immunostained with anti‐Ki‐67 (MIB‐1) antibody (M7240, 1:100; Dako, Glostrup, Denmark) using the standard procedure. The MIB‐1 labeling index was calculated as described.26
Drugs
Doxorubicin was obtained from Cell Signaling Technology (Tokyo, Japan), and thiostrepton was obtained from Millipore/EMD (Billerica, MA, USA). Both drugs were dissolved in DMSO obtained by Sigma‐Aldrich (St. Louis, MO, USA) and were used at the indicated concentrations.
Cytogenetic analysis
We carried out a cytogenetic analysis using TC616 cells. Metaphase cells were banded with Giemsa trypsin, and karyotypic descriptions were done according to the International System for Human Cytogenetic Nomenclature 2009.27
Pathologic studies of TC616 cell line
For routine light microscopy, TC616 cells were cultured on slides, fixed in methanol, and stained with H&E. The cells grown on slides were washed in PBS and fixed in cold acetone for 5 min. The cells were reacted with each of the primary antibodies for 1 h at room temperature. The primary antibody was monoclonal anti‐human α‐smooth muscle actin antibody (R&D Systems, St. Louis, MO, USA) diluted at 1:5000 and HHF35 (Enzo, Farmingdale, NY) diluted at 1:50. The bound antibodies were then visualized using the Dako EnVision System detection system. Paraffin sections from the original tumor were also examined using the same antibodies.
Small interfering RNA transfection
TC616 cells were transfected with On‐Targetplus Smart Pool siRNAs FOXM1 and On‐Targetplus Smart Pool siRNAs Control (Dharmacon, Brébières, France). On‐Targetplus Smart Pool siRNA FOXM1 contains four types of siRNA and target sequences were CCAACAAUGCUAAUAUUCA, CAUUGGACCAGGUGUUUAA, GCGCACGGCGGAAGAUGAA, and UCGAAAGACAUCUAUACGUG.
Transfections were undertaken using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocols. The introduction of the siRNA for FOXM1 was confirmed by quantitative RT‐PCR immunoblotting.
Cell viability
We assessed the viability of the TC616 cell line by performing an MTT assay using the Cell Counting Kit 8 (CCK‐8; Dojindo Molecular Technologies, Rockville, MD, USA) according to the manufacturer's instructions and as described.26 The absorbance at 450 nm was measured by a microplate reader (model 680; Bio‐Rad Laboratories, Hercules, CA, USA) by spectrophotometry at 450 nm. Assays were carried out in triplicate and were repeated at least three times in separate experiments.
TaqMan PCR to detect mRNA quantity of FOXM1
Quantitative RT‐PCR for FOXM1 was carried out and the results were analyzed using TaqMan assay reagents (FOXM1 Hs00170471_m1.; GAPDH Hs99999905_m1.; all from Applied Biosystems, Foster City, CA, USA) and an ABI Prism 7700 Sequence Detection system (Applied Biosystems). The PCR reaction was carried out according to the manufacturer's protocol. The obtained data were standardized using data of the housekeeping gene, GAPDH. The data were averaged from the values obtained in a triplicate reaction.
Western blot analysis
The TC616 cells were washed twice with ice‐cold PBS, scraped, and collected in a microcentrifuge tube. Whole cell lysates were prepared from the TC616 cell line. A total of 20 μg protein from each sample was used and incubated with anti‐FOXM1 (1:200 dilution) antibody (R&D Systems). Anti‐human actin mouse mAb (1:5000; Millipore) was used as a loading control. The subsequent procedures were carried out according to the manufacturer's protocol and as described.26
Chemosensitivity and proliferation assays of transfected cell lines
After 24‐h siRNA transfection, the transfected cells were seeded at 2000 cells per well in 96‐well plates. For the chemosensitivity assay, various concentrations of DOX were added to the medium after 12 h of incubation. After incubation for another 72 h, the number of viable cells in each well was measured. For the proliferation assay, the number of viable cells in each well was measured at 48, 72, and 96 h after transfection. Assays were carried out in triplicate and were repeated at least three times in separate experiments.
Drug treatment and cell proliferation assay
TC616 cells were plated on 96‐well plates at 2000 cells per well in serum‐containing growth medium. After a 12‐h incubation, the cells were treated with carrier alone (0.01% DMSO) as non‐treated controls and with DOX (5 ng/mL), thiostrepton (0.75 μM), or thiostrepton (0.75 μM) + DOX (5 ng/mL) for another 72 h. The resulting data are reported as the percentage of cell viability in comparison to that of the respective non‐treated control group (100%).
Statistical analysis
We used the χ2‐test and the t‐test as appropriate to evaluate associations between two variables. The Steel–Dwass multiple comparison test was applied to compare the data of more than two groups. The survival correlations are illustrated with Kaplan–Meier curves, and survival analyses were carried out using the log–rank test. In the multivariate analysis, a Cox proportional hazards model was used to independently examine factors. Two‐sided P‐values < 0.05 were considered significant.
Results
Prognostic significance of FOXM1 expression in leiomyosarcoma patients
Immunohistochemically, the positive expression of FOXM1 was recognized in 41 of the 123 LMS cases. Leiomyosarcoma cells showed nuclear staining for FOXM1 antibodies (Fig. 1a). In addition, the serial sections immunostained with anti‐Ki‐67 showed that some immunoreactive cells for FOXM1 antibody are also reactive for Ki‐67 (Fig. 1b).
Figure 1.
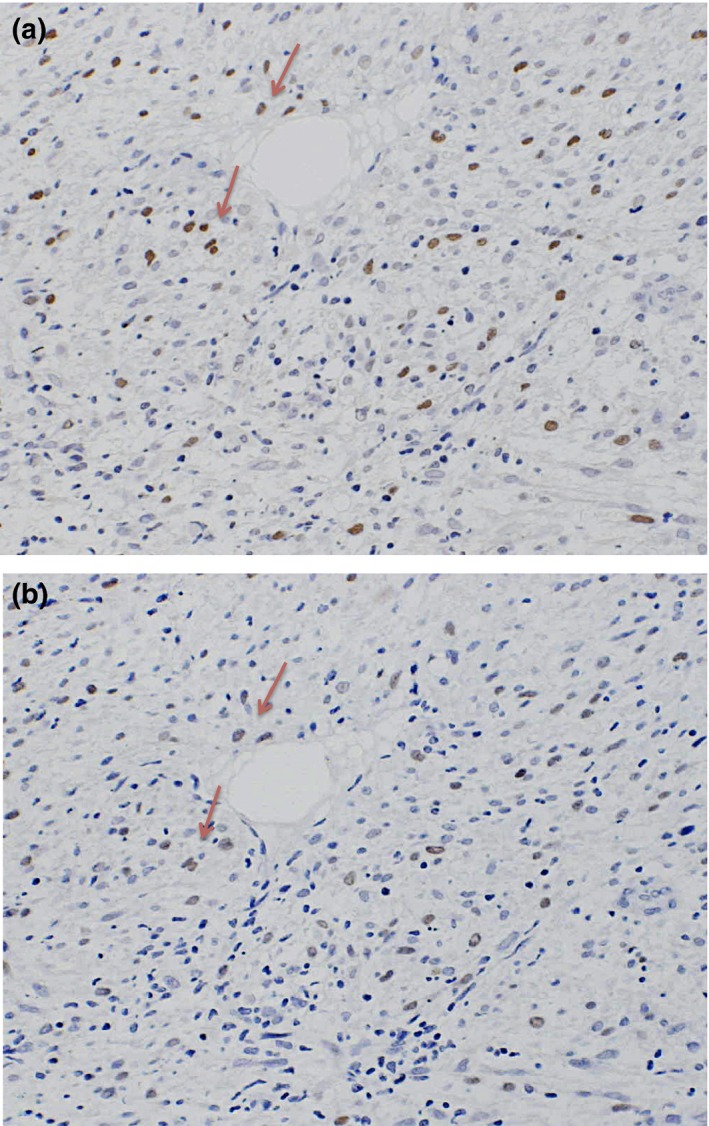
Serial sections of surgically resected soft tissue leiomyosarcoma tumors showing some immunoreactive cells for FOXM1 antibody are also reactive for Ki‐67 (red arrows). (a) Immunohistochemical results for FOXM1. Immunostaining for antibody was recognized in the nuclei. (b) Immunohistochemical results for Ki‐67.
The clinicopathologic data and the results of the survival analysis of all 123 patients are summarized in Table 1, as are the associations of clinicopathologic parameters with OS and EFS between groups. In the univariate analysis of prognostic factors, the expression of FOXM1 was significantly associated with OS (P = 0.0142) (Fig. 2A) and EFS (P = 0.0079) (Fig. 2b). Among the 28 patients treated with chemotherapy, the FOXM1 expression cases had poor prognoses in OS (P = 0.0189) (Fig. 2c). The following clinicopathologic variables were also associated with poor prognosis in OS: large tumor size (more than 5 cm), tumor depth (deep origin), tumor necrosis, high mitotic activity (mitotic count >10/high power field) and high FNCLCC histological grade (I vs. II; II vs. III).
Table 1.
Clinicopathologic parameters, FOXM1 expression, and results of survival analysis of patients with leiomyosarcoma of soft tissue (n = 123)
| Variable | No. of patients | Analyzed groups | P‐value | FOXM1 | |||
|---|---|---|---|---|---|---|---|
| OS | EFS | Positive | Negative | P‐value | |||
| Sex | |||||||
| Male | 61 | 19 | 42 | ||||
| Female | 62 | Male vs. female | 0.9862 | 0.33160 | 22 | 40 | 0.6009 |
| Age, years | |||||||
| <60 | 54 | 19 | 35 | ||||
| ≥60 | 69 | <60 vs. ≥60 | 0.3865 | 0.36210 | 22 | 47 | 0.7002 |
| Depth | |||||||
| Superficial | 35 | 12 | 23 | ||||
| Deep | 73 | Deep vs. superficial | 0.0308a | 0.11150 | 26 | 47 | 0.8921 |
| NA | 15 | ||||||
| Size, cm | |||||||
| <5 | 41 | 12 | 29 | ||||
| ≥5 | 70 | ≥5 vs. <5 | 0.0444a | 0.13690 | 27 | 43 | 0.3185 |
| NA | 12 | ||||||
| Necrosis | |||||||
| None | 61 | 21 | 40 | ||||
| <50% | 51 | Necrosis (+) vs. (−) | 0.0485a | 0.74050 | 16 | 35 | 0.4346 |
| ≥50% | 7 | 3 | 4 | ||||
| NA | 4 | ||||||
| Mitotic activity | |||||||
| ≤10/10HPF | 52 | 11 | 41 | ||||
| >10/10HPF | 68 | >10 vs. ≤10/10HPF | 0.0060a | 0.01700a | 30 | 38 | 0.0076a |
| NA | 3 | ||||||
| AJCC stage | |||||||
| I | 11 | I vs. II | 0.1155 | 0.19190 | 2 | 9 | 0.2664 |
| II | 73 | II vs. III | 0.2724 | 0.36110 | 25 | 48 | 0.2688 |
| III | 21 | III vs. IV | 0.9093 | – | 10 | 11 | 0.6579 |
| IV | 7 | 2 | 5 | ||||
| NA | 11 | ||||||
| FNCLCC | |||||||
| I | 33 | I vs. II | 0.0178a | 0.23540 | 8 | 25 | 0.7432 |
| II | 51 | II vs. III | 0.0045a | 0.01250a | 14 | 37 | 0.0165a |
| III | 36 | 17 | 19 | ||||
| NA | 3 | ||||||
| MIB‐1 labeling index (median 30/HPF) | |||||||
| Low (≤30) | 55 | High vs. low | 0.1032 | 0.04150a | 11 | 44 | 0.0070a |
| High (>30) | 58 | 29 | 29 | ||||
| NA | 10 | ||||||
| FOXM1 | |||||||
| Positive | 41 | ||||||
| Negative | 82 | Positive vs. negative | 0.0180a | 0.00496a | – | – | – |
P < 0.05 by log–rank test or χ2‐test. –, not caluculated. AJCC, American Joint Committee on Cancer; EFS, event‐free survival; FNCLCC, French Federation of Cancer Centers Sarcoma Group; HPF, high‐power field; NA, not available; OS, overall survival.
Figure 2.
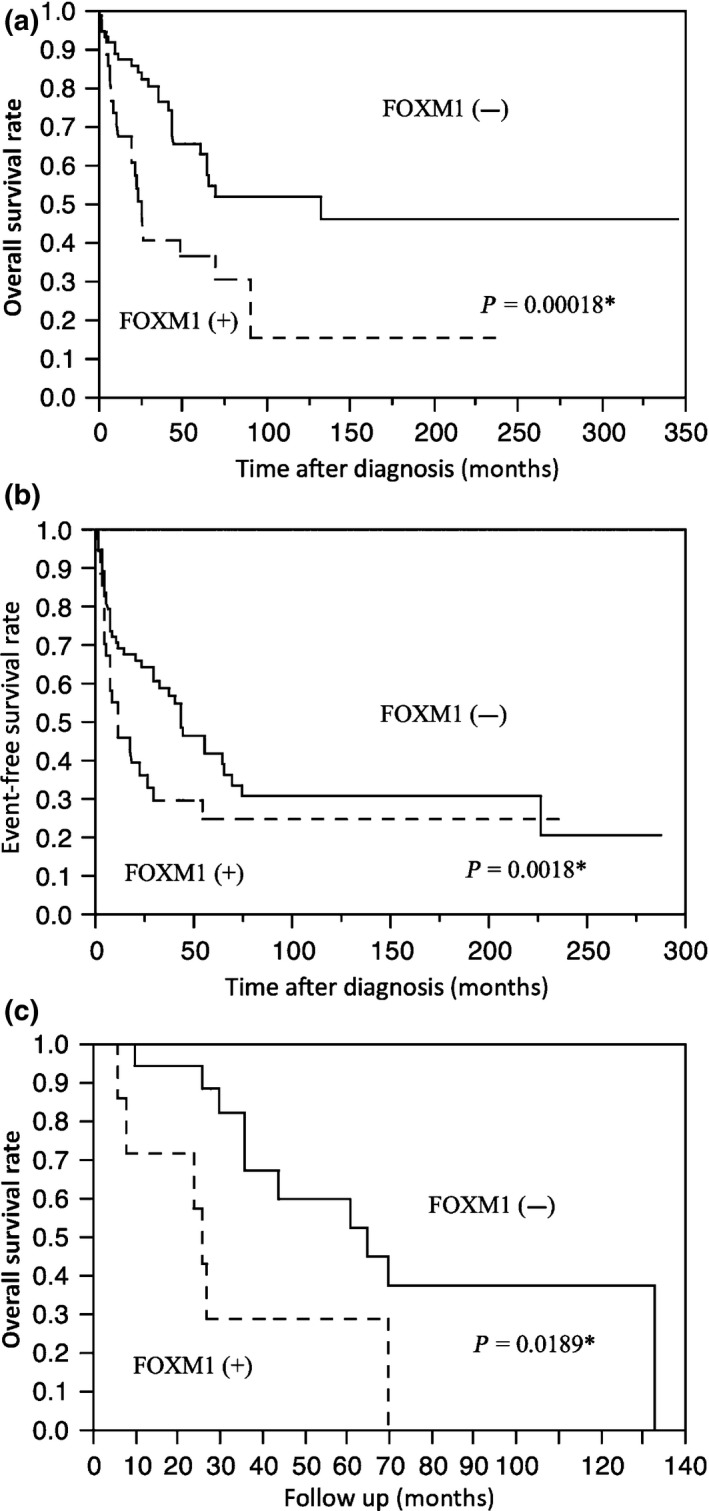
Kaplan–Meier survival curves for the overall survival (a) and event‐free survival (b) of patients with soft tissue leiomyosarcoma according to the results of the immunohistochemical study for FOXM1. (c) Overall survival for patients treated with chemotherapy. *P < 0.05 by log‐rank test.
The associations of clinicopathologic parameters with FOXM1 are also shown in Table 1. High mitotic activity, high MIB‐1 labeling index, and FNCLCC grade were shown to be correlated with the expression of FOXM1.
We also carried out a multivariate analysis for FOXM1 expression with clinicopathologic variables adjusted by tumor size, tumor depth, and FNCLCC grade, which were related to poor prognosis in the univariate analysis. We excluded the histological subtype, mitotic count, and necrosis, because the FNCLCC surgical stage is derived from them. The multivariate analysis revealed that FOXM1 expression and FNCLCC grade were significantly correlated with OS (Table 2).
Table 2.
Multivariate analysis of immunohistochemical and clinicopathological parameters
| Variable | P‐value (overall survival) |
|---|---|
| Tumor depth (deep vs. superficial) | 0.1856 |
| Tumor size (<5 vs. ≥5) | 0.4758 |
| FNCLCC grading (I vs. II, II vs III) | 0.0007a |
| FOXM1 (positive vs. negative) | 0.0162a |
P < 0.05 by Cox proportional hazards model.
Pathologic study of TC616 cell line
The original specimen of the TC616 cell line showed a proliferation of spindle‐ to oval‐shaped cells arranged in a fascicular pattern (Fig. 3a). The immunohistochemical study confirmed that the tumor cells of original specimen expressed FOXM1 (Fig. 3b). As assessed by light microscopy, the cells growing on slides were spindle‐shaped, round, or polygonal with abundant eosinophilic cytoplasm (Fig. 3c). The cells were immunoreactive for α‐smooth muscle actin (Fig. 3d), muscle‐specific actin (Fig. 3e), and FOXM1 (Fig. 3f) antibodies. These pathologic features indicated that TC616 cells maintained smooth muscle differentiation after 100 passages. Nuclear expression of FOXM1 in TC616 was also confirmed by immunohistochemical stain.
Figure 3.
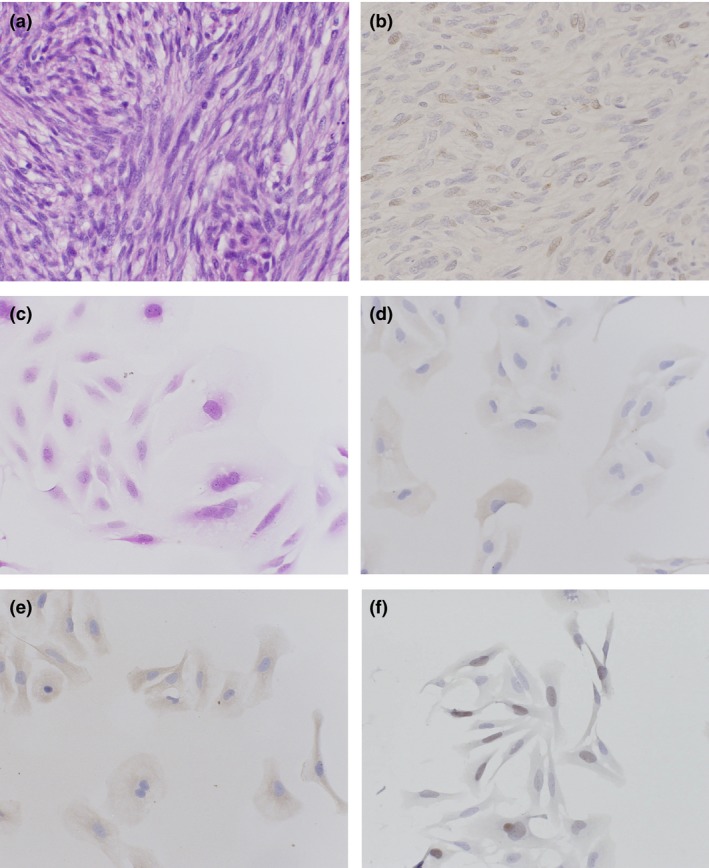
Hematoxylin–eosin stain (a) and immunohistochemical stain for FOXM1 (b) of the original tumor specimen of TC616 soft tissue leiomyosarcoma cells. Light microscopic findings of TC616 cells in vitro. The tumor cells were spindle, round, or polygonal in shape with oval nuclei and extension of slender cytoplasmic processes (c). Most TC616 cells showed immunopositive reactions for α‐smooth muscle actin (d), muscle‐specific actin (e), and FOXM1 (f) antibodies.
Cytogenic analysis of TC616 cells
A representative karyotype of TC616 cells is shown in Figure 4. The TC616 cells displayed a highly complex karyotype with numerous marker chromosomes. The composite karyotype was as follows:
Figure 4.

Representative G‐banded karyotype of a metaphase TC616 soft tissue leiomyosarcoma cell, including marker chromosomes. Arrows indicate the structural chromosome aberrations.
95‐117,‐X,‐XorY,add(X)(q13)×2,+1,add(1)(q32),del(1)(q11),der(1)add(1)(p13)del(1)(q23q25),‐2,add(3)(p21)×2,add(4)(p12),add(4)(p16),add(4)(q35),add(5)(p11),add(5)(p15.1),‐6‐6,‐7,add(7)(q32)×2,‐2,‐2,add(2)(p11.1),‐3,add(3)(q21),‐4,add(4)(q31.1),‐5,add(5)(q11.1),del(6)(q11)×2,del(7)(p11.1),del(7)(q11.1),der(7)add(7)(p22)add(7)(q22),‐8,add(9)(p11)×2,der(9)del(9)(p11)add(9)(q22),‐10,add(10)(p13),‐11,add(11)(q23),‐12,‐13,‐14,add(14)(p11.1),add(15)(p11.1),add(15)(p11.1),‐17,‐17,‐18,‐19,‐20,add(20)(q13.1),+add(21)(p11.1),‐22,‐22,+mar1,+mar2,+mar3,+mar4,+mar5,+mar6,+mar7,+mar8,+mar9,+mar10, +mar11,+mar12 [cp20].
Inhibition of FOXM1 by siRNA on TC616 cells
FOXM1 was knocked down in the TC616 cell line using siRNA. The interruption of FOXM1 was confirmed by Western blotting and RT‐PCR. (Fig. 5a,b). Reduced cell proliferation was recognized in the cell line after 96 h of transfection (t‐test, P = 0.0363) (Fig. 5c). Increased sensitivity for DOX at 10 ng/mL was observed in the TC616 cell line by FOXM1 interruption (Fig. 5d).
Figure 5.
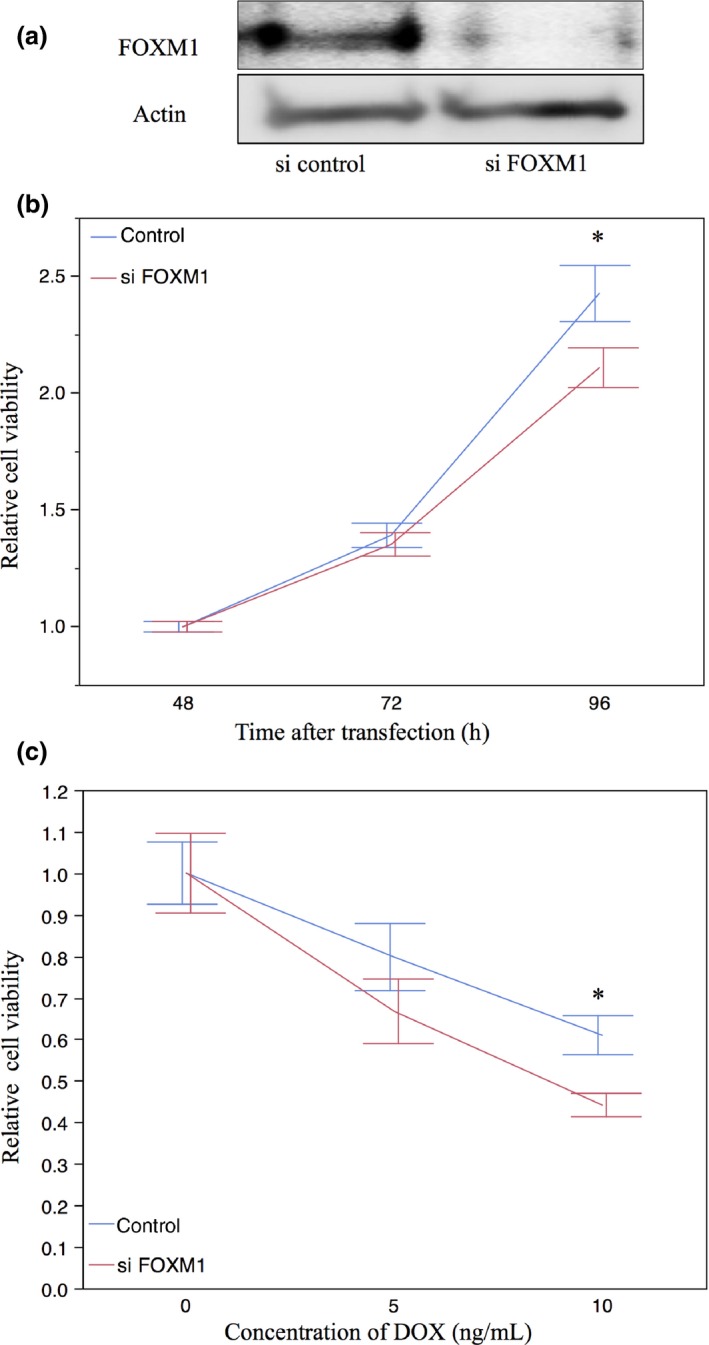
Proliferation and chemosensitivity assay results in TC616 soft tissue leiomyosarcoma cell lines with FOXM1. The cell lines were transduced with FOXM1 siRNA (si FOXM1) or a non‐targeting control (si control). (a) Western blot analysis showed that the cell lines transduced with FOXM1 had significantly reduced levels of FOXM1 protein at 48 h after transfection. (b) Real‐time quantitative PCR for FOXM1 showed a reduction in FOXM1 transcript at 24 h after transfection. (c) TC616 cells with FOXM1 siRNA compared to non‐targeting control. Significantly decreased proliferation was recognized in the TC616 cells at 96 h after transfection. (d) SiRNA targeting FoxM1 transfected cells had higher sensitivity for doxorubicin (DOX), compared with the control. Data are presented as mean ± SD for three independent experiments. *P < 0.05, t‐test.
Antitumor effect of thiostrepton in TC616 cells
Decreased FOXM1 expression compared to the untreated controls was revealed in the thiostrepton‐treated cells by Western blotting (Fig. 6a) and RT‐PCR (Fig. 6b). The FOXM1 inhibitor thiostrepton dose‐dependently inhibited the proliferation of TC616 cells (Fig. 6c). We also evaluated the effects of treatment with thiostrepton, DOX, and their combination on the proliferation of TC616 cells. We observed that the cells treated with the combination of both drugs showed lower proliferation compared to those treated with either drug individually (Steel–Dwass multiple comparison test, P < 0.05) (Fig. 6d).
Figure 6.
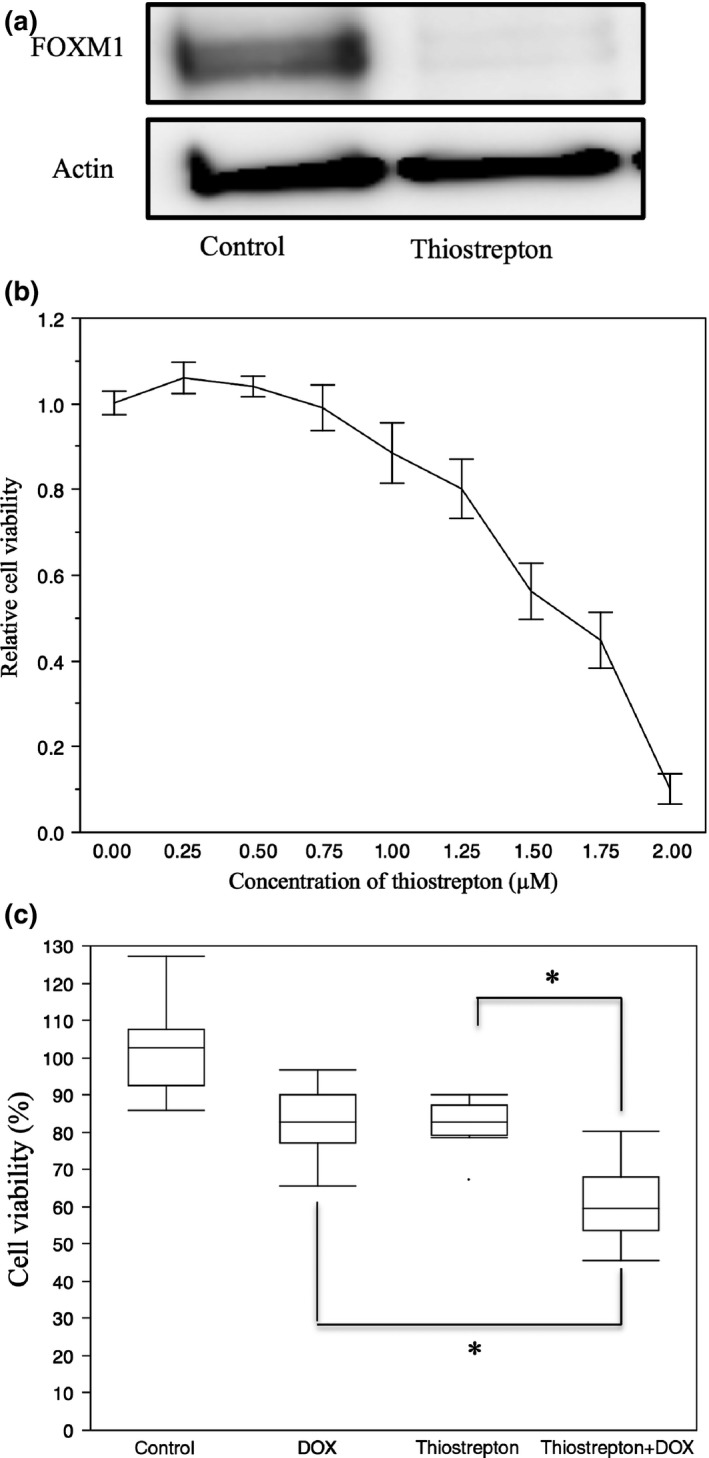
Thiostrepton reduced the FOXM1 expression in TC616 soft tissue leiomyosarcoma cells, resulting in diminished cell viability and increased chemosensitivity for doxorubicin (DOX). (a) TC616 cells treated with 0.75 μM thiostrepton for 48 h showed decreased FOXM1 protein on Western blots. (b) TC616 cells were treated with 0.75 μM thiostrepton for 48 h. Real‐time quantitative PCR showed a reduction of FOXM1 transcript. (c) Treatment of TC616 cells with increasing quantities of thiostrepton for 72 h resulted in reduced numbers of viable cells compared to diluent controls. (d) Proliferation of TC616 cells treated with 0.75 μM thiostrepton, 5 ng/mL DOX, or their combination. The cells treated with both drugs showed significantly decreased cell viability compared to cells treated with each drug individually. *P < 0.05, Steel–Dwass multiple comparison test.
Discussion
The expression of FOXM1 in clinical specimens has been reported to be an adverse prognostic factor8, 12, 13, 14, 16, 17, 21 in many malignancies and is regarded as a possible target of molecular therapy. In the present study, FOXM1 expression was correlated with poor prognosis for OS among LMS patients in the univariate analysis. In the multivariate analysis adjusted by depth, tumor size, and FNCLCC grade, FOXM1 expression was an independent prognostic factor for OS. Among the clinicopathologic factors, high FOXM1 expression was correlated with high mitotic activity, high MIB‐1 labeling index, and high FNCLCC grade. These histological factors are also associated with poor prognosis, and systemic therapy is more likely to be needed in histologically aggressive LMS. FOXM1 expression was revealed to also be correlated with poor prognosis for OS among LMS patients treated with chemotherapy. These findings indicate that FOXM1 is a reliable biomarker for adverse prognosis in LMS patients.
On the basis of these pathologic findings, we have established a new LMS cell line, TC616, which originated from FOXM1‐expressing LMS tissue. We carried out a proliferation and cell viability assay for FOXM1 interruption by using siRNA and the FOXM1 inhibitor thiostrepton in TC616 cells.
In the TC616 cell line, the immunohistochemical staining for smooth muscle markers and morphological characteristics was maintained after 100 passages. Many cases of LMS have highly complex karyotypes lacking specific structural or numerical aberrations.2 As expected, the TC616 cells had complex karyotypes with a number of numerical and structural alterations, including marker chromosomes.
Interruption of FOXM1, achieved by using siRNA, caused a reduction in cell proliferation. The interruption of FOXM1 decreases cell proliferation through an inhibition of cell‐cycle progression in many cancer cell lines.18, 23 FOXM1 is one of a few genes shown to be upregulated during early cancer development, and the involvement of FOXM1 in the onset of tumorigenesis may be related mainly to its role in cell‐cycle progression and proliferation.28, 29, 30, 31 FOXM1 interruption caused decreasing viability treated with DOX in TC616 cells. Doxorubicin treatment of cancer cells created double‐stranded DNA breaks; FOXM1 regulates DNA repair genes and is involved in chemoresistance through a DNA repair pathway.32
We evaluated the antitumor effect of the FOXM1 inhibitor thiostrepton.33 Thiostrepton reduced the number of viable TC616 cells in a dose‐dependent manner. Thiostrepton also reduced the expression of FOXM1 protein in this cell line. The cytotoxicity might not only manifest through the inhibition of FOXM1; the mechanisms of FOXM1 interruption by thiostrepton have been proposed to be through the direct binding of FOXM133 and also its activity as a proteasome inhibitor.34 However, little is known about the efficacy of proteasome inhibitors in soft tissue sarcomas.35
Doxorubicin is routinely available for sarcoma treatment in many countries. The first‐line chemotherapy for advanced, metastatic, or non‐resectable soft tissue sarcoma is typically based on DOX as a single agent or in combination with a second drug such as ifosfamide.36 The DOX treatment of cancer cells creates double‐stranded DNA breaks, and DNA repair genes are induced to rescue the cells from the DNA damage. FOXM1 regulates DNA repair genes (XRCC1 and BRCA2), and is involved in chemoresistance through a DNA repair pathway.37 The interruption of FOXM1 expression in breast cancer cells sensitized the cells to DOX.37
In the present study, the LMS cell line TC616 treated with the combination of thiostrepton and DOX showed lower cellular proliferation than the cells treated with either drug individually and FOXM1 interruption achieved by using siRNA caused improved chemosensitivity in TC616. Thiostrepton has the potential to be an agent for treating LMS with high FOXM1 expression, based on its ability to bind FOXM1 directly.
The findings we obtained using clinical samples and in vitro experiments are consistent with previously reported mechanisms of FOXM1 in other malignancies. This therapeutic intervention has minimal toxicity in normal tissue, because of low FOXM1 expression in normal adult tissue. The efficacy of proteasome inhibitors in LMS has not yet been elucidated, but the single use of thiostrepton or a combination of thiostrepton and conventional chemotherapy may be a suitable treatment for LMS cases with overexpression of FOXM1.
In summary, our present findings clarified that FOXM1 inhibition is a candidate treatment option for soft tissue leiomyosarcoma, based on our clinicopathologic assessment and an in vitro study using thiostrepton and siRNA on an LMS cell line. Further in vivo and in vitro investigations are warranted to evaluate the efficacy of the FOXM1 inhibitor thiostrepton alone and in combination with other agents.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
The authors thank KN International for revising the English used in this article. This work was supported in part by a Grant‐in‐Aid for Scientific Research (B) (25293088) from the Japan Society for the Promotion of Science (to YO).
Cancer Sci 107 (2016) 95–102
Funding Information
Japan Society for the Promotion of Science.
References
- 1. Fletcher CD, Bridge AJ, Hogendoom PC et al World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon, France: IARC Press; 2013: 213–5. [Google Scholar]
- 2. Wang R, Lu YJ, Fisher C et al Characterization of chromosome aberrations associated with soft‐tissue leiomyosarcomas by twenty‐ four‐color karyotyping and comparative genomic hybridization analysis. Genes Chromosom Cancer 2001; 31: 54–64. [DOI] [PubMed] [Google Scholar]
- 3. Svarvar K, Böhling T, Berlin Ö et al Clinical course of nonvisceral soft tissue leiomyosarcoma in 225 patients from the Scandinavian sarcoma group. Cancer 2007; 109: 282–91. [DOI] [PubMed] [Google Scholar]
- 4. Korver W, Roose J, Clevers H. The winged‐helix transcription factor Trident is expressed in cycling cells. Nucleic Acids Res 1997; 25: 1715–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yao KM, Sha M, Lu Z et al Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J Biol Chem 1997; 272: 19827–36. [DOI] [PubMed] [Google Scholar]
- 6. Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res 2005; 65: 5181–9. [DOI] [PubMed] [Google Scholar]
- 7. Koo C‐Y, Muir KW, Lam EWF. Review FOXM1: from cancer initiation to progression and treatment. Biochim Biophys Acta 2012; 1819: 28–37. [DOI] [PubMed] [Google Scholar]
- 8. Yang DK, Son CH, Lee SK et al Forkhead box M1 expression in pulmonary squamous cell carcinoma: correlation with clinicopathologic features and its prognostic significance. Hum Pathol 2009; 40: 464–70. [DOI] [PubMed] [Google Scholar]
- 9. Ha SY, Lee CH, Chang HK et al Differential expression of forkhead box M1 and its downstream cyclin‐dependent kinase inhibitors p27 (kip1) and p21 (waf1/cip1) in the diagnosis of pulmonary neuroendocrine tumours. Histopathology 2012; 60(5): 731–9. [DOI] [PubMed] [Google Scholar]
- 10. Teh MT, Wong ST, Neill GW et al FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res 2002; 62: 4773–80. [PubMed] [Google Scholar]
- 11. Ahmad A, Wang Z, Kong D et al FoxM1 down‐regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra‐cellular matrix degrading factors. Breast Cancer Res Treat 2010; 122: 337–46. [DOI] [PubMed] [Google Scholar]
- 12. Huang C, Qiu Z, Wang L et al A novel FoxM1‐caveolin signaling pathway promotes pancreatic cancer invasion and metastasis. Cancer Res 2012; 72: 655–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu XY, Zhu ZM, Chen LB et al FOXM1 expression correlates with tumor invasion and a poor prognosis of colorectal cancer. Acta Histochem 2012; 114: 755–62. [DOI] [PubMed] [Google Scholar]
- 14. Priller M, Pöschl J, Abrão L et al Expression of FoxM1 is required for the proliferation of medulloblastoma cells and indicates worse survival of patients. Clin Cancer Res 2011; 17: 6791–801. [DOI] [PubMed] [Google Scholar]
- 15. Kim IM, Ackerson T, Ramakrishna S et al The Forkhead box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res 2006; 66: 2153–61. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Wen L, Zhao SH et al FoxM1 expression is significantly associated with cisplatin‐based chemotherapy resistance and poor prognosis in advanced non‐small cell lung cancer patients. Lung Cancer 2013; 79: 173–9. [DOI] [PubMed] [Google Scholar]
- 17. Li X, Qi W, Yao R et al Overexpressed transcription factor FOXM1 is a potential diagnostic and adverse prognostic factor in postoperational gastric cancer patients. Clin Transl Oncol 2014; 16: 307–14. [DOI] [PubMed] [Google Scholar]
- 18. Nakamura S, Hirano I, Okinaka K et al The OXM1 transcriptional factor promotes the proliferation of leukemia cells through modulation of cell cycle progression in acute myeloid leukemia. Carcinogenesis 2010; 31: 2012–21. [DOI] [PubMed] [Google Scholar]
- 19. Wan X, Yeung C, Kim SY et al Identification of FoxM1/Bub1b signaling pathway as a required component for growth and survival of rhabdomyosarcoma. Cancer Res 2012; 72: 5889–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Christensen L, Joo J, Lee S et al FOXM1 is an oncogenic mediator in Ewing sarcoma. PLoS ONE 2013; 8: e54556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yu J, Deshmukh H, Payton JE et al Array‐based comparative genomic hybridization identifies CDK4 and FOXM1 alterations as independent predictors of survival in malignant peripheral nerve sheath tumor. Clin Cancer Res 2011; 17: 1924–34. [DOI] [PubMed] [Google Scholar]
- 22. Wang IC, Chen YJ, Hughes D et al Forkhead box M1 regulates the transcriptional network of genes. essential for mitotic progression and genes encoding the SCF (Skp2‐Cks1) ubiquitin ligase. Mol Biol Cell 2005; 25: 10875–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grant GD, Brooks L 3rd, Zhang X et al Identification of cell cycle‐regulated genes periodically expressed in U2OS cells and their regulation by FOXM1 and E2F transcription factors. Mol Biol Cell 2013; 24(23): 3634–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halasi M, Gartel AL. Suppression of FOXM1 sensitizes human cancer cells to cell death induced by DNA‐damage. PLoS ONE 2012; 7(2): e31761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edge SB, Byrd DR, Compton CC et al AJCC Cancer Staging Manual, 7th edn St. Louis, MO: Springer, 2010. [Google Scholar]
- 26. Endo M, Yamamoto H, Setsu N et al Prognostic significance of AKT/mTOR and MAPK pathways and antitumor effect of mTOR inhibitor in NF1‐related and sporadic malignant peripheral nerve sheath tumors. Clin Cancer Res 2012; 15: 19. [DOI] [PubMed] [Google Scholar]
- 27. Shaffer LG, Slovak ML, Campbell LJ, ISCN . An International System for Human Cytogenetic Nomenclature. Basel: Karger, 2009. [Google Scholar]
- 28. Uddin S, Ahmed M, Hussain A et al Genome‐wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol 2011; 178: 537–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Costa RH, Kalinichenko VV, Major ML, Raychaudhuri P. New and unexpect‐ ed: forkhead meets ARF. Curr Opin Genet Dev 2005; 15: 42–8. [DOI] [PubMed] [Google Scholar]
- 30. Francis RE, Myatt SS, Krol J et al FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol 2009; 35: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kretschmer C, Sterner‐Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer 2011; 10: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol 2007; 27: 1007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem 2011; 3: 725–31. [DOI] [PubMed] [Google Scholar]
- 34. Bhat UG, Halasi M, Gartel AL. FoxM1 is a general target for proteasome inhibitors. PLoS ONE 2009; 4: e6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maki RG, Kraft AS, Scheu K et al A multicenter Phase II study of bortezomib in recurrent or metastatic sarcomas. Cancer 2005; 103: 1431–8. [DOI] [PubMed] [Google Scholar]
- 36. Schöffski P, Cornillie J, Wozniak A, Li H, Hompes D. Soft tissue sarcoma: an update on systemic treatment options for patients with advanced disease. Oncol Res Treat 2014; 37: 355–62. [DOI] [PubMed] [Google Scholar]
- 37. Park YY, Jung SY, Jennings NB et al FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis 2012; 33(10): 1843–53. [DOI] [PMC free article] [PubMed] [Google Scholar]


