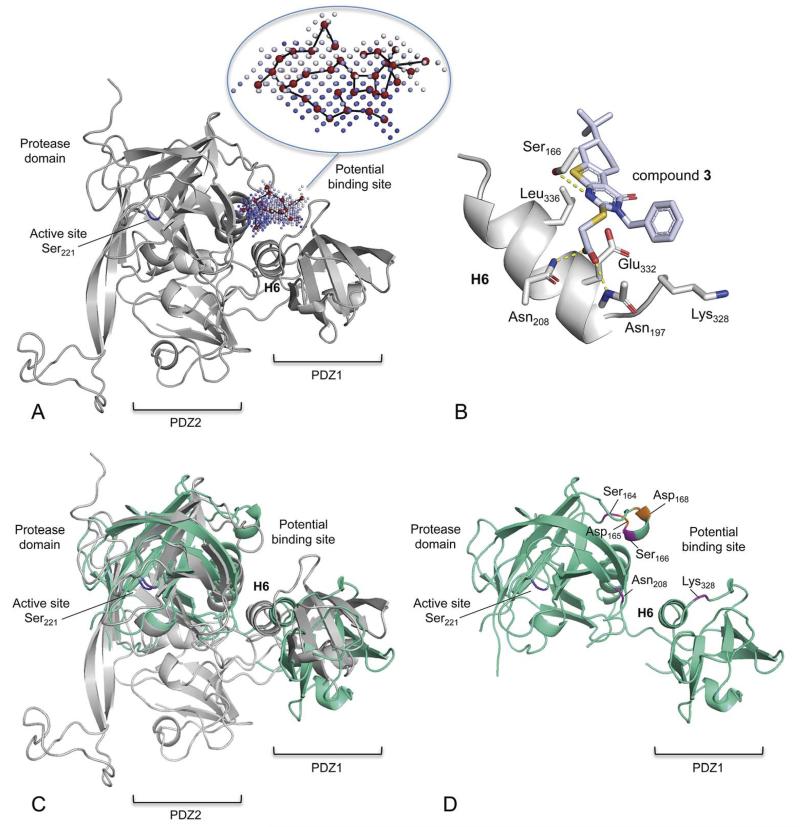
Fig. 3.
Structural model of H. pylori HtrA and a potential binding mode of compound 3. (A) Comparative (“homology”) protein model of HpHtrA. The enzyme contains a protease domain and two PDZ domains (PDZ1, PDZ2). The potential allosteric pocket that was used for virtual screening is located between the protease domain and the PDZ1 domain. This pocket lies distant to the active site (catalytic Ser221) and is flanked by helix 6 (H6) and two loops. The pocket graph (red vertices) computed by the PoLiMorph software is shown inside the allosteric pocket that was extracted by the PocketPicker tool. The intensity of the blue colour of the pocket grid points correlates with their buriedness. (B) Computed docking pose of compound 3. Potential hydrogen-bond interactions are indicated by yellow dotted lines. (C) Structural alignment of the homology model (gray) and a preliminary X-ray structure of HpHtrA (green). (D) X-ray structural model of HpHtrA with the mutated residues highlighted.