Figure 4.
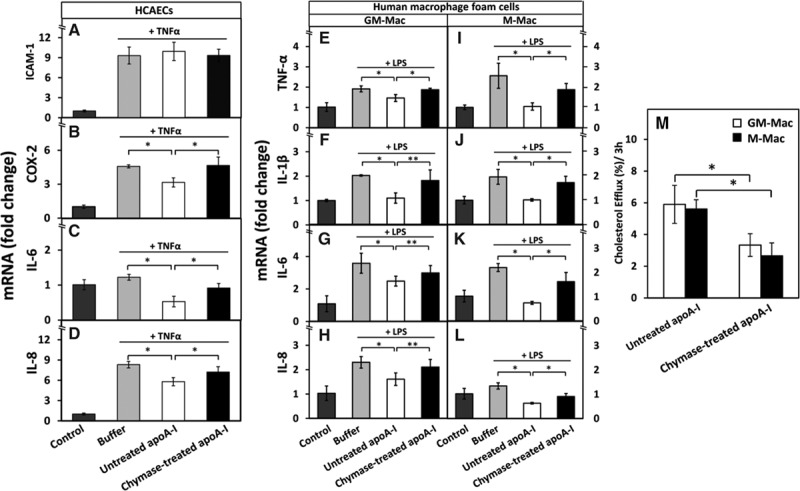
Proteolysis of apolipoprotein A-I (apoA-I) reduces its ability to downregulate the expression of proinflammatory genes in human coronary artery endothelial cells (HCAECs) and in human GM-Mac and M-Mac macrophage subpopulations and to promote cholesterol efflux from the macrophages. A-D, HCAECs were preincubated with untreated or chymase-treated apoA-I and then activated with tumor necrosis factor-α (TNF-α), as described in Figure 1. Nonactivated cells (control) and TNF-α-activated cells preincubated in only medium (buffer) acted as a reference. The mRNA levels of intercellular adhesion molecule-1 (ICAM-1), cyclooxygenase-2 (COX-2), interleukin (IL)-6, and IL-8 in HCAECs were measured by quantitative real-time polymerase chain reaction (qRT-PCR) and expressed as fold changes relative to the control cells. Data represent the means±SD from 3 independent experiments performed in duplicate. *P<0.01. E-L, Human monocyte-derived macrophages were differentiated in the presence of GM-CSF (granulocyte-macrophage colony-stimulating factor) or M-CSF (macrophage colony-stimulating factor) into GM-Mac and M-Mac subtypes, respectively, and then converted into foam cells by incubation with acetyl low density lipoprotein (LDL). The cells were then incubated for 3 h with untreated or chymase-treated apoA-I (50 μg/mL, each), washed, and activated for 3 h with lipopolysaccharide (LPS). Nonactivated cells (control) and LPS-activated cells preincubated in only medium (buffer) acted as references. LPS-induced mRNA levels of TNF-α, IL-1β, IL-6, and IL-8 in GM-Mac and M-Mac foam cells were evaluated by qRT-PCR and expressed as fold changes relative to the control cells. *P<0.01; **P<0.05. M, GM-Mac- and M-Mac-derived foam cells were incubated with untreated or chymase-treated apoA-I (50 μg/mL, each) for 3 h, the media were collected, centrifuged to remove cellular debris, and the radioactivity of each supernatant was determined by liquid scintillation counting. Cells were solubilized, and radioactivity was determined in the cell lysates. Cholesterol efflux was calculated as dpmmedium/(dpmcells+dpmmedium)×100. Data represent the means±SD from triplicate wells and are representative of 2 independent experiments. *P<0.01.
