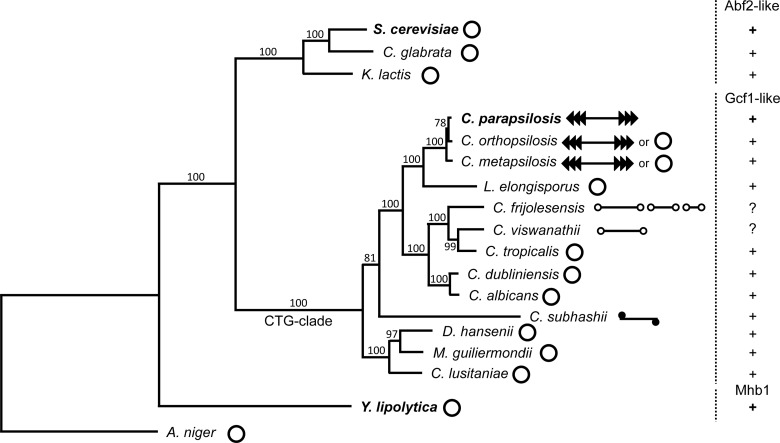
Figure 6. Phylogenetic tree illustrating the distribution of various types of mtHMG proteins and variability of the form of mitochondrial genomes in selected yeast species.
The phylogeny was calculated from concatenated multiple sequence alignments of mtDNA-encoded proteins (i.e. Atp6-8-9-Cob-Cox1-2-3-Nad1-2-3-4-4L-5-6-Rps3) by the maximum likelihood algorithm and LG (Le-Gascuel) amino acids substitution model implemented in the PhyML program [80]. Bootstrap values (out of 100 replicates) are shown above the corresponding branches. Aspergillus niger from the subphylum Pezizomycotina was used as an outgroup. Mitochondrial genome forms were classified as described previously [76] and are illustrated by pictograms (open circles–circular; lines with open circles at the ends–linear with terminal hairpins (i.e. type 1 linear and multipartite type 1 linear); lines with series of arrowheads–linear with array of tandem repeats (i.e. type 2 linear); line with closed circle at the ends–linear with a protein covalently bound to 5′ termini (i.e. type 3 linear). The species whose mtHMG proteins were investigated in the present study are shown in bold.