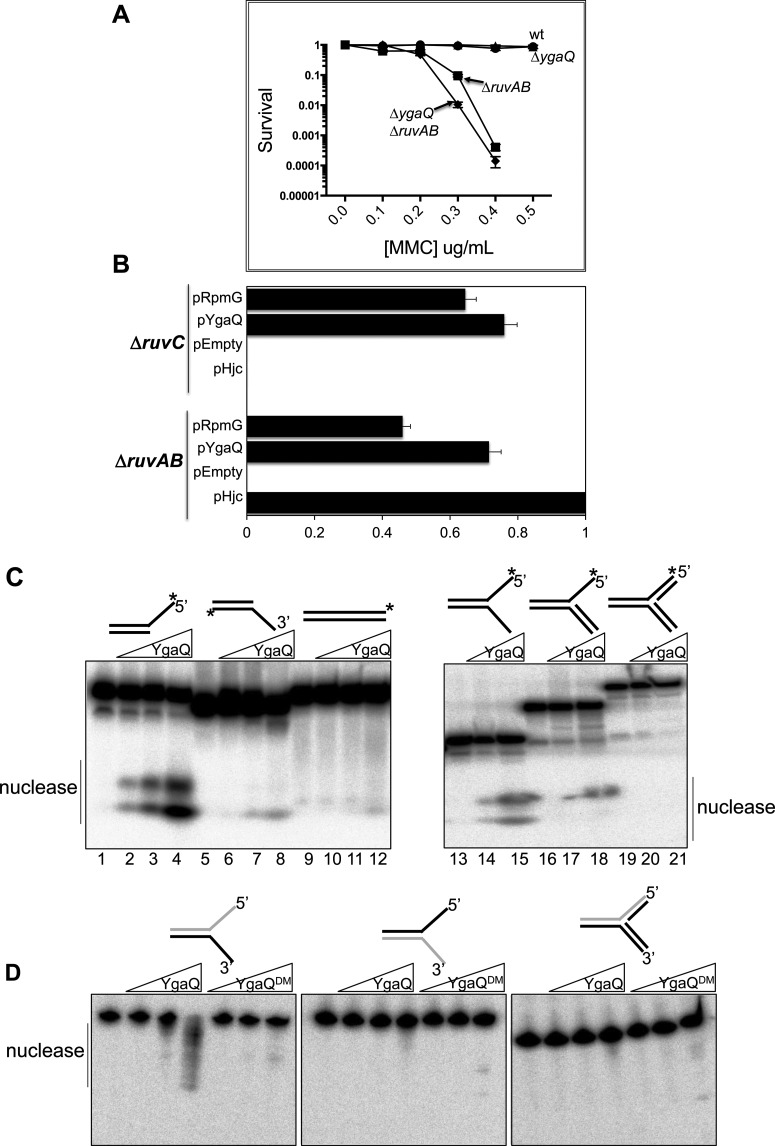
Figure 3. YgaQ is a nuclease that acts independently of Holliday junction processing by RuvABC.
(A) Graph ‘killing curves’ comparing strains ΔygaQ, ΔruvAB and ΔruvAB ΔygaQ for MMC sensitivity in viability spot tests plotted as a function of MMC concentration as indicated. The assays were done in triplicate with bars representing standard error. (B) Graph showing survival of ASKA plasmids expressing YgaQ (pYgaQ) or RpmG (pRpmG) compared with the positive control pHjc and corresponding empty ASKA plasmid vector. Assays were done twice and standard error from the mean is given as bars. (C) Non-denaturing TBE acrylamide gel for analysis of products from mixing YgaQ with DNA substrates as indicated. YgaQ was used at 0, 2.5, 25 and 250 nM (lanes 1–12) or 0, 25 and 250 nM (lanes 13–21) in reactions containing 0.6 nM of DNA that was 32P 5′-end-labelled as indicated with (*). (D) Urea denaturing TBE acrylamide gels for analysis of products from mixing YgaQ with forked DNA as indicated; in each substrate the strand presented in grey is labelled at its 5′ end. YgaQ and YgaQDM mutant proteins were each used at 0, 2.5, 25 or 250 nM in reactions containing 0.6 nM of DNA.