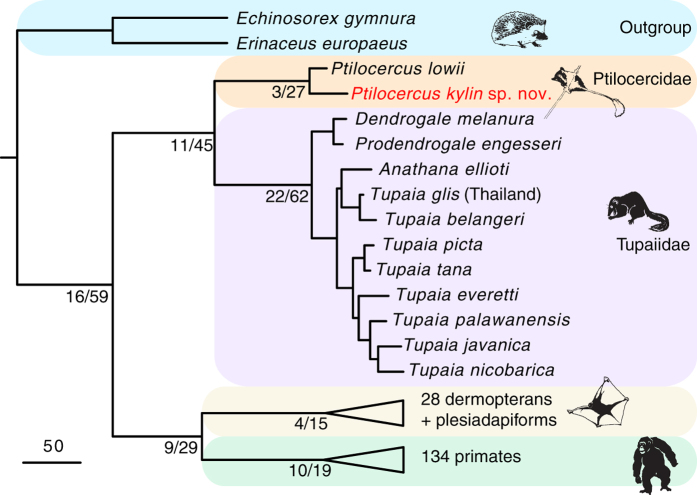
Figure 3. Summary phylogeny of treeshrews.
Parsimony analysis is based on a data matrix including 1199 morphological characters and 658 molecular characters of long and short interspersed nuclear elements scored for 130 fossil and 47 living taxa. The topology of extant treeshrews, flying lemurs and primates used as a backbone constraint or “molecular scaffold” is based on a gene supermatrix (ref. 50). Numbers before the slashes at the internodes are the absolute Bremer Support values; numbers after the slashes are Relative Bremer Support values. The strict consensus of the unconstrained analysis is provided in the Supplementary Information. The topologies of treeshrews in both the constrained and unconstrained analysis are identical. Scale bar equals 50 character changes.