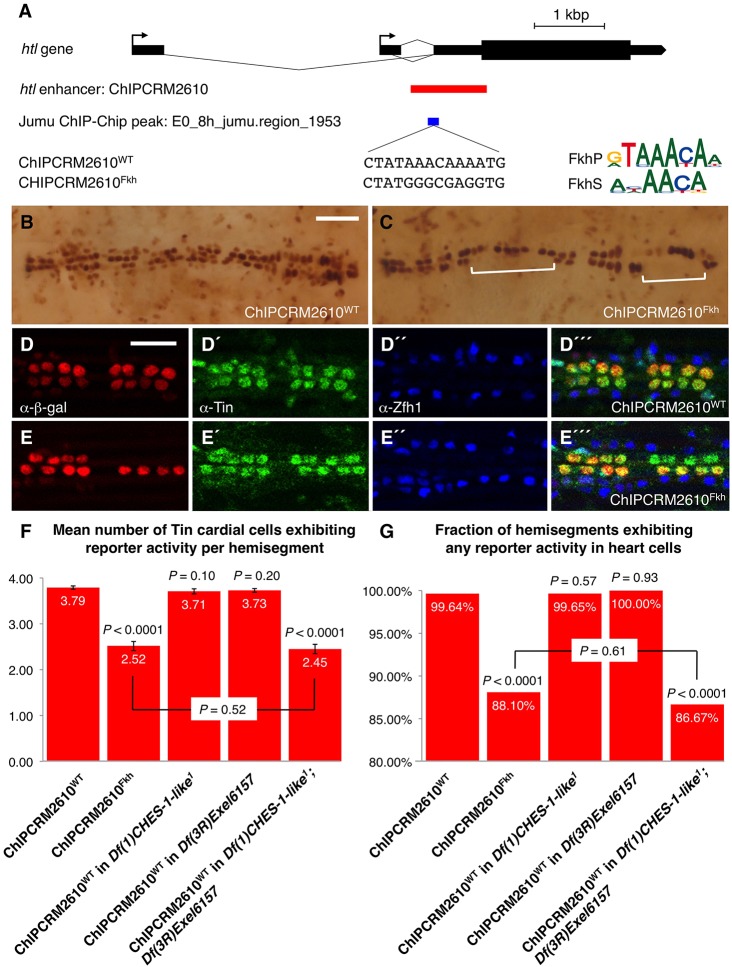
Fig. 4.
CHES-1-like and Jumu are activators of a htl cardiac enhancer. (A) Relative positions of the htl gene, the htl enhancer ChIPCRM2610, the Jumu ChIP peak, and the Fkh TF binding sequence. Logo representations of the PWMs of Fkh primary (FkhP) and secondary (FkhS) TF binding motifs obtained from protein-binding microarrays are shown, as are the Fkh binding sequences in the wild-type (ChIPCRM2610WT) and the mutated (ChIPCRM2610Fkh) enhancer. (B,C) Representative Stage 16 hearts showing β-galactosidase reporter activity driven by the wild-type (B) and mutant (C) enhancers. Square brackets indicate entire hemisegments lacking reporter activity in the case of the mutated enhancer (C). Scale bar: 50 μm. (D-E‴) Enhancer activity at single-cell resolution. The posterior-most four CCs in a hemisegment are marked by Tin expression (green), and the PCs are marked by Zfh1 expression (blue). (D-D‴) Reporter activity (red) driven by the wild-type enhancer is located primarily in the Tin-expressing CCs. (E-E‴) Mutating the Fkh binding site results in significant reduction of reporter activity. Scale bar: 25 μm. (F,G) Quantification and significance of the effects of mutating the Fkh binding site in the ChIPCRM2610 htl enhancer, and the effects of loss of function of CHES-1-like, jumu, or both CHES-1-like and jumu on the wild-type ChIPCRM2610 htl enhancer activity. The P-value over each column indicates the significance of the difference in reporter activity between the relevant enhancer genotype combination and the wild-type ChIPCRM2610WT enhancer in wild-type embryos. Error bars in F indicate 95% confidence intervals.