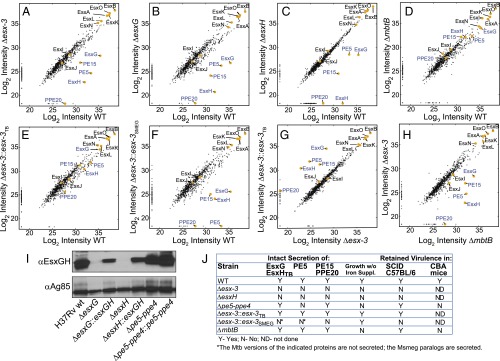
Fig. 6.
Proteomics MS analysis of CF reveals codependent secretion of PE5 with EsxG–EsxH and identifies PE15/PPE20 as ESX-3 substrates. (A–H) Scatterplots show log2-transformed LFQ intensity values of proteins identified in the CF with the following strain comparisons: H37Rv vs. Δesx-3 (mc27844) (A); H37Rv vs. ΔesxG (mc27845) (B); H37Rv vs. ΔesxH (mc27846) (C); H37Rv vs. ΔmbtB (mc27850) (D); H37Rv vs. Δesx-3::esx-3TB (mc27827) (E); H37Rv vs. Δesx-3::esx-3SMEG (mc27828) (F); Δesx-3 vs. Δesx-3::esx-3TB (G); and ΔmbtB vs. Δesx-3 (H). Values for proteins of interest (Esx proteins and select PE–PPE proteins) are in yellow and are labeled; proteins implicated as ESX-3 substrates are indicated by blue text. Trace amounts of PE5, EsxG, and EsxH were detected in the Δesx-3 samples because of carryover from other samples; when the columns were washed extensively between runs, PE5, EsxG, and EsxH were absent from Δesx-3, as anticipated (Fig. S5C). See Fig. S6 for lists of proteins that are present at significantly different levels for the various comparisons and Dataset S2 for lists of all proteins detected in the secretome analysis. (I) Western blot analysis of CF from strains grown in chelated Sauton’s medium using the EsxG–EsxH antibody. Ag85B served as a loading control. (J) Summary of secretome findings, in vitro growth requirements, and in vivo growth characteristics for the various strains.