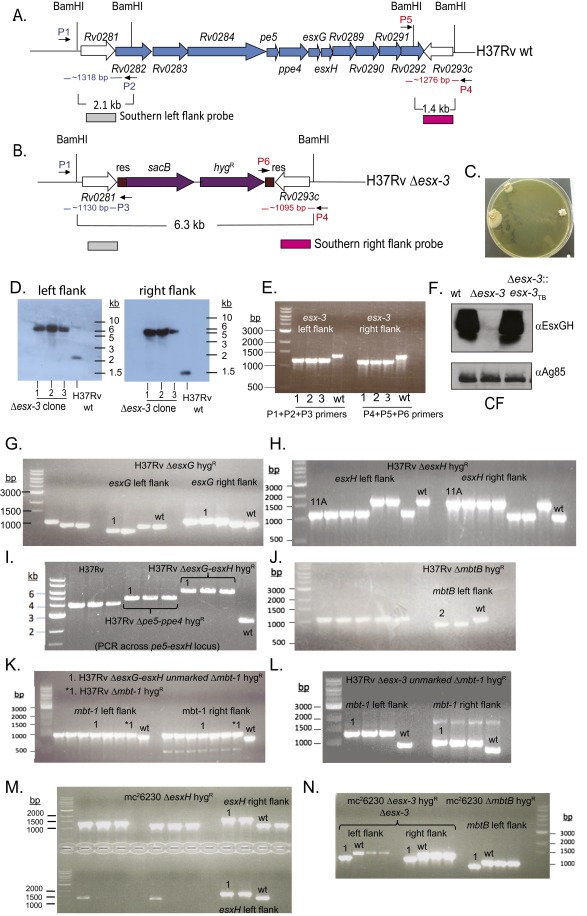
Fig. S1.
Confirmation of mutations in the esx-3 and mbt-1 regions. (A and B) The esx-3 locus (Rv0282–Rv0292) is shown. ORFs removed in the Δesx-3 deletion strain are shown in blue (A). The resulting genomic organization including the sacB-hygR cassette (purple) flanked by γδ-resolvase (res) sites (brown) is shown in B. The location of primers used to verify the esx-3 deletion by PCR in E is indicated in blue (left flank; P1, P2, P3) and red (right flank; P4, P5, P6). (C) Δesx-3 colonies were obtained on plates supplemented with 100 μM hemin and 0.05% Tween 80. Colonies were initially visible after ∼4 wk of incubation and were photographed at ∼7 wk of incubation. (D) Southern blotting was used to confirm deletion of esx-3 from three transductants (1–3), using the probes indicated in A and B. Genomic DNA prepared from H37Rv Δesx-3 transductants (1–3) and H37Rv WT was digested with BamHI. Blots were probed with sequences immediately flanking the deleted esx-3 region at the locations indicated in A and B. Probes were amplified from H37Rv gDNA using the primer pairs ΔRv0282LL+ΔRv0282LR and ΔRv0292RL+ΔRv0292RR, respectively (Dataset S1). Relevant BamHI restriction sites are shown in A and B. The probes for the left flank or the right flank each hybridized to an ∼6.3-kb band in the mutants. In contrast, the WT control yielded the expected band sizes of ∼2.1 and ∼1.4 kb, respectively. (F) Western blotting of CF demonstrated that Δesx-3 failed to secrete EsxG and EsxH. Antigen 85B served as a loading control. (E and G–N) PCR analysis was performed to confirm the mutant genotypes in the H37Rv background for Δesx-3 (mc27788) (E); ΔesxG (mc27789) (G); ΔesxH (mc27790) (H); Δpe5–ppe4 (mc27792) and ΔesxG–esxH (mc27791) (I); ΔmbtB (mc27808) (J); ΔesxG–esxH Δmbt-1 (mc27851) and Δmbt-1 (mc27809) (K); Δesx3 Δmtb-1 (mc27852) (L) and in the mc26230 strain background ΔesxH (mc27819) (M); and Δesx-3 (mc27818) and ΔmbtB (mc27820) (N). Screening primers yielded distinct band sizes for mutant and WT strains and are listed in Dataset S1. The H37Rv WT and mc26230 parental strains are indicated, as are the clones selected for study (numbers above lanes, indicating clone numbers in the laboratory collection). Primers amplify the left and right flanking regions of the targeted genes, except for the Δpe5–ppe4 and ΔesxG–esxH mutants shown in I, where the primers are upstream and downstream of the deleted region. Left- and right-flanking PCRs also were performed subsequently for these mutants and yielded the expected product sizes.