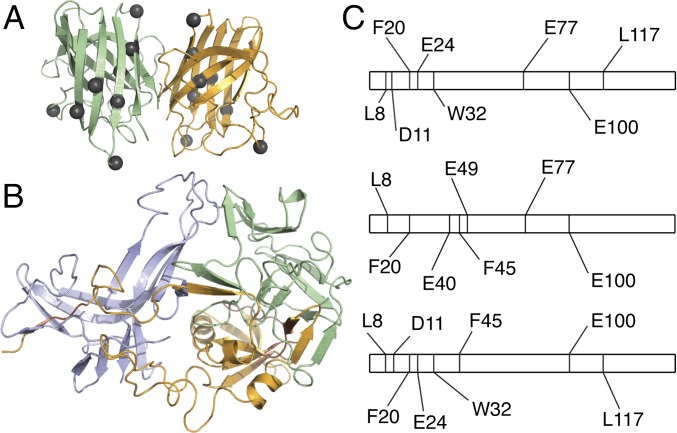
Fig. 2.
Hybrid experimental/computational method leads to a model of metastable SOD1 trimer. (A) SOD1 trimer proteolytic cut sites (gray spheres, shown here on the native dimer structure) in the dimer interface and secondary structural elements suggest significant structural differences between native and trimeric structures. (B) Structural model of the metastable SOD1 trimer obtained using limited proteolysis data as constraints in several rounds of coarse-grained and atomistic DMD simulations (Fig. S2). (C) Representation of the SOD1 linear sequence, residues 1–153. Proteolytic cut sites (vertical lines) determined in limited proteolysis experiments differ significantly in SOD1 monomer (Top), dimer (Middle), and trimer (Bottom), supporting structural rearrangement during the aggregation process and SOD1 trimer formation.