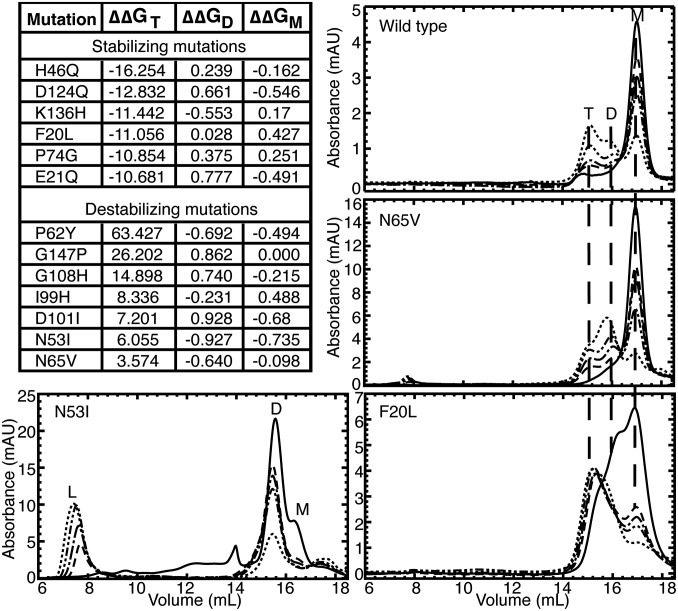
Fig. 3.
Designed mutations verify the model and demonstrate control of SOD1 aggregation. Mutations to trimer interface residues designed to stabilize or destabilize SOD1 trimer but having no effect on SOD1 monomer or dimer, are shown with ΔΔGmut for the trimer, native dimer, and native monomer structures. Aggregation time courses were measured for each mutant after incubation at physiological concentration (30 μM) and 37 °C for for 0 h (solid line), 2 h (− −), 4 h (− − •), 8 h (− • •), or 24 h (• •). The aggregation of apo-WT SOD1 is shown for comparison, with trimer (T), dimer (D), monomer (M), and large aggregate (L) peaks labeled when present. Vertical dashed lines between panels aid comparison of trimer, dimer, and monomer peaks between mutants. We find that the N65V mutation, predicted to be destabilizing to the SOD1 trimer, results in a smaller population of trimer than WT, shifting the SOD1 population toward dimer and monomer formations. The N53I mutation, also predicted to destabilize SOD1 trimer, results in no detectable SOD1 trimer but, instead, increased populations of large aggregates. The F20L mutation, predicted to stabilize SOD1 trimer, promotes trimer formation early in the aggregation process and maintains high levels of trimer throughout the experiment. Additional aggregation time courses can be found in Fig. S5. mAU, milli-absorbance units.