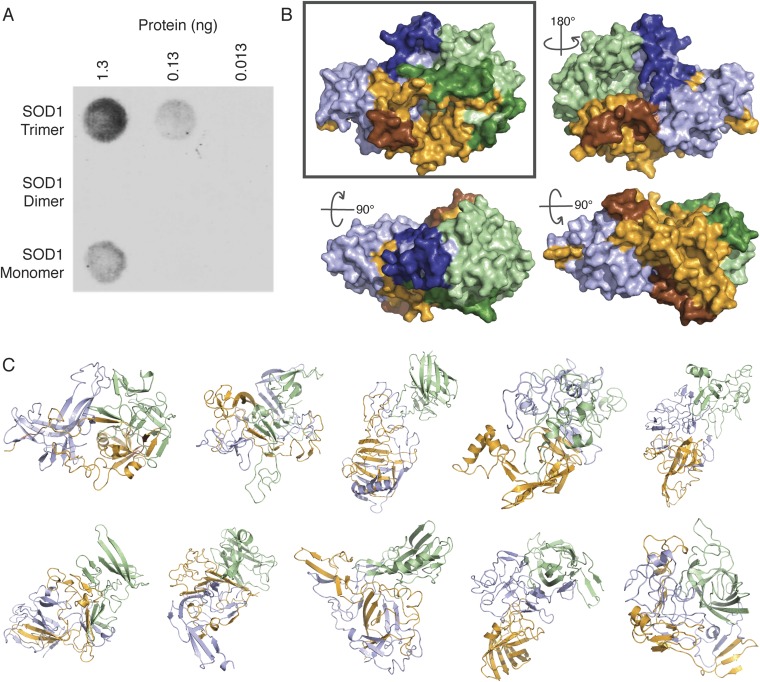
Fig. S4.
Toxic epitope exposed on SOD1 trimer surface. Ensemble of SOD1 trimer models. (A) Conformational Ab C4F6, which binds several disease-associated species of SOD1 (16), selectively binds to WT SOD1 trimer at various concentrations, but does not bind to native WT type SOD1 dimer and exhibits minimal binding to WT SOD1 monomer. (B) Recently identified epitope of the C4F6 Ab (16, 18) is exposed on the surface of our SOD1 trimer model. Individual monomers are depicted in pale green, bright orange, and light blue; residues comprising the C4F6 epitope are highlighted in darker colors: forest, brown, and deep blue, respectively. Rotation angles describe the transformation from the “front” (Upper Left), such that the structures represent the “back” (Upper Right), “top” (Lower Left), and “bottom” (Lower Right). The C4F6 Ab has been shown previously to bind WT and disease-linked mutant SOD1 trimer at physiological pH (15). In addition to experimental data from limited proteolysis, the exposure of the C4F6 epitope further verifies our model, as well as providing support for the toxicity of SOD1 trimer. (C) Replicate SOD1 trimer models resulting from 10 independent repetitions of our protocol (Fig. S3) feature significant variation in tertiary and quaternary structure. Models vary in the degree of domain swapping, as well as in the amount of native tertiary structure maintained in each monomer. We note that despite differences in tertiary and quaternary structure of the 10 SOD1 trimer models, a high level of consensus on the identity of residues involved in trimeric interface interactions is maintained, with at least 77% and as much as 94% identity in interface residue identity between any two models.