Abstract
Cell-membrane lipid composition can greatly influence biophysical properties of cell membranes, affecting various cellular functions. We previously showed that lipid synthesis becomes altered in the membranes of resistant breast cancer cells (MCF-7/ADR); they form a more rigid, hydrophobic lipid monolayer than do sensitive cell membranes (MCF-7). These changes in membrane lipids of resistant cells, attributed to epigenetic aberration, significantly affected drug transport and endocytic function, thus impacting the efficacy of anticancer drugs. The present study’s objective was to determine the effects of the epigenetic drug 5-aza-2′-deoxycytidine (DAC), delivered in sustained-release nanogels (DAC-NGs), on the composition and biophysical properties of membrane lipids of resistant cells. Resistant and sensitive cells were treated with DAC in solution (DAC-sol) or DAC-NGs, and cell-membrane lipids were isolated and analyzed for lipid composition and biophysical properties. In resistant cells, we found increased formation of Cholesterol-Sphingomyelin (CHOL-SM) rafts with culturing time, whereas DAC treatment reduced their formation. In general, the effect of DAC-NGs was greater in changing the lipid composition than with DAC-sol. DAC treatment also caused a rise in levels of certain phospholipids and neutral lipids known to increase membrane fluidity while reducing the levels of certain lipids known to increase membrane rigidity. Isotherm data showed increased lipid membrane fluidity following DAC treatment, attributed to decrease levels of CHOL-SM rafts (lamellar beta [Lβ] structures or ordered gel) and a corresponding increase in lipids that form lamellar alpha structures (Lα, liquid crystalline phase). Sensitive cells showed marginal or insignificant changes in lipid profile following DAC-treatment, suggesting that epigenetic changes affecting lipid biosynthesis are more specific to resistant cells. Since membrane fluidity plays a major role in drug transport and endocytic function, treatment of resistant cells with epigenetic drugs with altered lipid profile could facilitate anticancer drug transport to overcome acquired drug resistance in a combination therapy.
Introduction
Cell-membrane lipids play a crucial role in such cellular functions as cell signaling,1 regulation of membrane trafficking,2 transmembrane protein function (e.g., P-glycoprotein),3 apoptotic pathways,4 drug transport,5 and endocytosis.6 The role that cell-membrane characteristics such as lipid composition and membrane biophysical properties (rigidity/fluidity) play in cell-membrane trafficking and the membrane’s role in different diseases is only beginning to be known.7
Multidrug resistance is a major clinical issue in anticancer drug therapy. Changes in lipid composition and the biophysical properties of membrane lipids have been implicated in drug resistance in several cell lines.8–12 We recently reviewed the role of membrane lipids in cancer progression and drug resistance.13 In our previous studies, we demonstrated that composition and biophysical characteristics of membrane lipids of drug-resistant breast cancer cells (MCF-7/ADR) are significantly different than that of drug-sensitive cells (MCF-7). The lipids of resistant cells contained hydrophobic lipids and formed a more rigid monolayer than the lipids of sensitive cells.5 The rigid nature of the resistant cell membrane impaired endocytic function, thus influencing the effect of Doxil, a commercial liposomal formulation of doxorubicin, whereas hydrophobic lipids partitioned doxorubicin in the membrane, thus influencing its transport and hence efficacy.6 Similarly, several other drug-resistant cell lines have been reported to show changes in lipid composition and biophysical properties of membrane lipids, but the role of these factors in drug transport and efficacy has not been investigated in depth.14–15
A common feature in acquired drug resistance is high levels of sphingomyelin (SM) and/or cholesterol (CHOL); these lipids can form “raft” regions that increase lipid packing density and lower fluidity, thus restricting drug transport.5, 14–15 We and others have previously shown that higher SM levels in resistant breast cancer cells is due to methylation silencing of the gene for the enzyme sphingomyelinase (SMase); hence, the basal activity of SMase (which hydrolyzes SM) is lower in resistant cells than in sensitive cells.6, 16 In this study, we analyzed changes in lipid composition and the impact of such changes on biophysical properties of cell-membrane lipids with sustained DAC effect in nanogels (NGs) (DAC-NGs) in resistant (MCF-7/ADR) cells. Our hypothesis was that DAC-NGs due to better and sustained delivery than with DAC-sol would effectively alter lipid biosynthesis, which would also influence the biophysical characteristics of resistant cell-membrane lipids. Our data show greater effect of DAC-NGs than DAC-sol in depleting CHOL-SM rafts from resistant cells whereas the effect of DAC treatment was marginal on the lipid profile of sensitive cells.
Experimental
Materials
Sphingomyelin (SM) was purchased from Avanti Polar Lipid (Alabaster, AL). Ethylenediaminetetraacetic acid (EDTA), tris(hydroxymethyl)aminomethane (Tris), ammonium hydroxide, diethyl ether, toluene, dimethyl sulfoxide, n-hexane, benzene, N-isopropylacrylamide, vinyl pyrrolidone, sodium dodecylsulfate, sodium acrylate, N,N′-cystamine bisacrylamide (S-S, a disulfide crosslinker), ammonium persulfate, maleic anhydride and 5-aza-2′-deoxycytidine (decitabine; DAC), cholesterol (CHOL) were purchased from Sigma-Aldrich (St. Louis, MO). Doxorubicin hydrochloride was purchased from Drug Source Co. LLC (Westchester, IL). Poly(ethylene glycol) (M.W. ~5000) was purchased from Polysciences, Inc. (Warrington, PA). Hydrochloric acid, sodium chloride, isopropanol, petroleum ether, glacial (water-free) acetic acid, ethanolic phosphomolybdic acid, copper sulfate pentahydrate, 98% sulfuric acid, 85% orthophosphoric acid of reagent grade, and chloroform, methanol, and isopropanol of high-performance liquid chromatography grade were purchased from Fisher Scientific (Pittsburgh, PA).
Nanogel synthesis and DAC loading
Nanogels (NGs) based on N-isopropylacrylamide (700 mg) were synthesized using an emulsion polymerization technique in the presence of poly(ethylene glycol)-maleic anhydride polymer (100 mg) using vinyl pyrrolidone (100 mg) as co-monomer, sodium dodecylsulfate (200 mg) as surfactant, ammonium persulfate as initiator, and N,N′-cystamine bisacrylamide as crosslinker, at 70 °C for 6 h, as per our previously described method.17 DAC was loaded into previously formed NGs by adding DAC (300 μL, 8.1 mg/mL dimethyl sulfoxide) to NGs (30 mg) dispersed in 6 mL of water, followed by 3 h stirring at 4 °C. The NG suspension was dialyzed for 30 min in a cold room to remove the unentrapped DAC; the above time for dialysis was determined sufficient to remove free DAC. The DAC-loaded NGs were then lyophilized over two days at −48 °C, 3.5 Pa, using FreeZone 4.5 (Labconco Corp., Kansas City, MO). The various conditions for synthesis of NGs and DAC loading were optimized in our previous study.17 The amount of DAC loading in NGs was determined using high-performance liquid chromatography (HPLC) and NGs were characterized for mean particle size in water using dynamic light scattering at a scattering angle of 90° at 25 °C and zeta potential in the phase analysis mode and the current mode at a scattering angle of −14° (NICOMP™ 380 ZLS, Particle Sizing Systems, Santa Barbara, CA). DAC-NGs were typically 244 nm in hydrodynamic diameter (polydispersity index =0.11) with zeta potential of −19 ± 1.0 mV, and DAC loading of 6.4 ± 0.4% w/w.17 Sustained antiproliferative effect of DAC-NGs as compared to DAC-sol in various cancer cell lines including in MCF-7/ADR was shown in our previous study.17 Release of DAC from NGs in vitro could not be carried out because of its instability in an aqueous medium at physiological temperature.18
Cell Culture
Doxorubicin-resistant breast cancer (MCF-7/ADR) cells were grown in Dulbecco’s modified Eagle’s (DMEM) supplemented with 15% fetal bovine serum (Gibco BRL, Grand Island, NY) and 100 μg/mL penicillin G and 100 μg/mL streptomycin at 37 °C in a humidified and 5% CO2 atmosphere. Sensitive cells were cultured in Eagle’s minimum essential medium supplemented with Earle’s salts, L-glutamine, 100 μg/mL streptomycin, 10% fetal bovine serum. These conditions were determined optimal for normal growth of sensitive and resistant cells. The difference in amount of fetal bovine serum used for culturing resistant and sensitive cells are not expected to have any effect on their cell membrane lipid composition. Cell culture media used to culture both cell lines were obtained from the Central Cell Services’ Media Laboratory of our institution. Resistant cells were cultured with media containing 100 ng/mL of doxorubicin after every two passages to maintain drug resistance.
DAC Treatment
Both resistant and sensitive cells were cultured in 150 × 25 mm culture dish (BD Biosciences, San Jose, CA) at a seeding density of 5 × 106 cells per dish in 25 mL of cell culture media. The cells were allowed to attach for 24 h prior to receiving different treatments. Cells were either treated with a single dose of DAC-sol or DAC-NGs (equivalent to 50 ng/mL DAC). This dose of DAC was selected because it was found to be nontoxic to these cells in our previous study.6 Untreated group either received no treatment or control NGs (without DAC). No further doses of DAC were added or medium was changed. The cells were harvested at each time post-treatment (1, 5 and 8 d) first by washing cells with ice-cold D-PBS and then scraping them into 10 mL of cold-sterile water using a cell scraper (Corning, Inc., Lowell, MA). Water was used for scrapping cells to avoid the effect of salts in the buffer in isolating and purification of lipids. Typically, cell suspensions from six dishes were combined for each treatment and time point to obtain enough lipid for lipid analysis and isotherm. The cells were centrifuged at 1,300 rpm for 7 min at 4 °C (Sorvall Legend RT centrifuge, Thermo Fisher Scientific, Waltham, MA). The resulting cell pellet was suspended in 3 mL of sterile water. The suspension was lyophilized for 48 h as described above. The lyophilized cell mass was stored at −80 °C until used for lipid extraction. Each experiment was carried out in triplicate for each treatment and time point.
Lipid Extraction
Lipids from the lyophilized cell mass were extracted by using a modified Bligh and Dyer method, as described in our previous study.5 Briefly, the lyophilized cell mass was dispersed in N2-purged deionized water (3 mL) to which a 10.2 mL mixture of methanol:chloroform:1 M HCl (23:10:1 v/v/v) was added. The mass was vortexed for 1 min and the container kept in an ice bath for 15 min to obtain a monophasic cell mass suspension. To the monophasic cell mass suspension, 3 mL chloroform and 3 mL of 0.1 M HCl were added and the materials vortexed to obtain a biphasic cell mass suspension. The biphasic cell mass suspension was separated by centrifugation at 3,500 rpm at 0 °C for 5 min (Sorvall Legend RT centrifuge). The organic phase from the bottom was collected carefully using a glass syringe and placed in a 30 mL glass vial (Fisher Scientific). The organic phase was then mixed with 3 mL of sodium chloride-Tris-EDTA buffer mixture (0.1 M NaCl, 0.1 M EDTA, and 0.05 M Tris buffer; pH 8.2). The extraction protocol was repeated from the remaining aqueous phase to ensure the complete recovery of the lipids. The above two buffer-organic phase extracts were combined, vortexed, and centrifuged as above to separate the organic phase-containing lipids. The organic phase with lipids was mixed with isopropanol (organic phase:isopropanol (15:1 v/v) and stored at −20 °C until used for protein separation, as described below.
Phospholipid Extraction
Phospholipids were extracted from the total lipid mixture by means of solid phase extraction.19 In brief, the total lipid extract was dissolved in 1.0 mL of n-hexane and transferred into a preconditioned silica gel-bonded column (Supelclean LC-Si, 6 mL volume, 1 g sorbent; Supelco (Sigma-Aldrich), Bellefonte, PA). The silica gel-bonded column was preconditioned by rinsing the packed column with 10 mL of n-hexane. First, fatty acids from the lipid extracts were removed by rinsing the column with 10 mL of a diethyl ether:n-hexane mixture (1:4 v/v). Then hydrocarbons, CHOL esters and triacylglycerols were removed by eluting the column with 18 mL of a 2-propanol:chloroform mixture (1:2 v/v). Finally, phospholipids were eluted with 10 mL of methanol followed by 10 mL of a chloroform:methanol:water mixture (3:5:2 v/v/v). The phospholipid fraction collected was evaporated to dryness in a rotary evaporator, as described above. The residue was dissolved in chloroform (200 μL) for phospholipid quantification, as described below.
Lipid Analysis
CHOL and SM levels in lipids from resistant cells were quantified by HPLC using an evaporative light-scattering detector (Shimadzu Scientific Instruments, Inc., Columbia, MD) and N2 as a nebulizing gas.19 The following method parameters were used: an HPLC column (100 mm × 4.6 mm, macropore size 2.1 μm and mesopore size 13 nm (Chromolith Performance Si HPLC column, EMD Millipore, an affiliate of Merck KGaA, Darmstadt, Germany); column temperature, −25 °C; mobile phase A, methanol:chloroform:ammonia solution (19.5:80:0.5 v/v/v); mobile phase B, methanol:chloroform:triethylamine:water (25.5:69.5:0.49:4.4, v/v/v/v); flow rate, 1 mL/min; injection volume, 10 μL; nebulizer temperature, 50 °C; detector gain, 6. The method parameters were optimized by using a mixture of phospholipids for HPLC analysis. Separation of SM and CHOL from phospholipids was achieved by the following gradient elution method, in which solvent B was increased from 0% to 40% within 5 min after which B was kept constant at 40% for 2 min. B was then increased from 40% to 100% within the next 6 min, after which B was kept constant for 7 min. B was decreased from 100% to 0% within 5 min; a post-run of 5 min was done to equilibrate the column before the next injection. The quantification of CHOL and SM was carried out by a pre-drawn calibration curve.
In addition, lipids were analyzed by high-performance thin-layer chromatography (HPTLC) technique since it is difficult to analyze all the lipids present in membrane lipid extracts using HPLC, either due to problems in identifying different peaks, separation of peaks, and/or non-availability of different lipid standards for identification and quantification lipids in a sample. HPTLC is semi-quantitative but can identify different lipids based on their retention time. In a typical experiment, HPTLC plates (10 cm×10 cm, Sigma-Aldrich) were preconditioned by drying at 140 °C for 30 min. To the HPTLC plate, a 5 μL aliquot of lipid extract solution (5 mg/mL) was added at a distance of 1 cm from the bottom of the plate. The mobile phase was allowed to run in a trough chamber containing 50 mL of the mobile phase for a distance of 8 cm. Post run, plates were dried under N2 gas for 15 min. Different mobile phases and staining procedures were used to separate and identify neutral lipids, phospholipids and ceramides from the lipid extracts. For instance, phospholipids were separated by using a mobile phase consisting of a mixture of methanol:chloroform:water:ammonia (75:120:6:2 v/v/v/v) and identified by immersing them in a copper sulfate solution for 5 s, followed by heating at 140 °C for 30 min. Neutral lipids were separated by a mobile phase consisting of diethyl ether:petroleum ether:glacial acetic acid (20:80:1 v/v/v) and marked by using 5% ethanolic phosphomolybdic acid solution, followed by heating at 140 °C for 30 min. Ceramides were separated by a mobile phase consisting of methanol:toluene (3:7 v/v) and stained with iodine vapor.
Biophysical Characterization
A Langmuir balance (Minimicro, KSV NIMA [Biolin Scientific], Helsinki, Finland) was used to study the surface pressure – area (π – A) isotherms of lipid mixtures. In general, to obtain the π – A isotherm, 2.5 μL of a chloroform:methanol (4:1 v/v) solution of cell-membrane lipids extracted as above (5 mg/mL) was added dropwise (~0.5 μL) onto the Dulbecco’s phosphate-buffered saline (D-PBS) subphase (pH 7.4) in the trough using a Hamilton digital microsyringe (Hamilton Co., Reno, NV). After waiting for 10 min to allow the chloroform to evaporate, the barriers were compressed at the rate of 5 mm/min until the collapse of the membrane.
The changes in biophysical characteristics of resistant cell-membrane lipids with culturing time and following treatment with DAC were assessed using the following parameters: A0′ (the extrapolated % trough area), membrane fluidity, πmax (maximum surface pressure before the collapse of the monolayer) and % trough area at πmax. A0′ is the minimum area required by all the lipids in the trough to form closest packing in the monolayer at the solid condensed/liquid condensed (SC/LC) phase. A0′ is arrived at by measuring the % trough area that corresponds to the slope of the SC/LC phase at zero surface pressure. A0′ is inversely proportional to lipid packing density. The membrane fluidity is arrived at by calculating surface compression modulus, which characterizes the lipid monolayer’s resistance to compression at the interface, from the surface pressure (SP) – area (π – A) isotherm data, as described in our previous studies.5 The membrane fluidity is inversely proportional to compression modulus. A high πmax indicates a high degree of adsorption of the lipids at the interface. Therefore, similarity in πmax and % trough area at πmax can be used as a measure to assess similarity of composition between two lipid mixtures.
Statistical Analysis
Data are expressed as mean ± standard error of mean. Statistical analyses were performed using one-way analysis of variance.
Results
Analysis of cholesterol and sphingomyelin levels with culturing time and following treatment with DAC
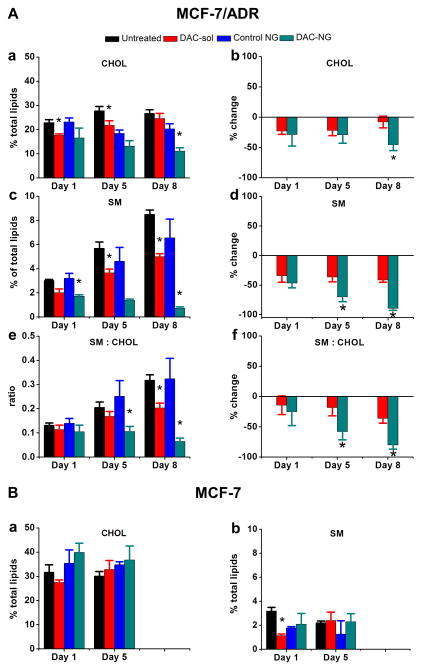
Analysis of resistant cell-membrane lipids by HPLC showed significant increase in SM content but marginal change in CHOL content with culturing time (Figure 1Aa and c). Greater increase in SM levels than CHOL levels with culturing time is clearly evident when the data are plotted as a ratio of SM:CHOL levels (Figure 1Ae).
Figure 1. Analysis of levels of CHOL and SM with culturing time and following treatment with DAC.
Resistant (A) and sensitive cells (B) were treated with either DAC in solution (DAC-sol) or in nanogels (DAC-NGs) (50 ng/mL). Lipids were extracted at different time points post treatment and analyzed for CHOL and SM. CHOL and SM levels from resistant cells were analyzed using HPLC whereas those in sensitive cells were analyzed using HPTLC. Data were normalized to the respective controls, and changes are shown as percent reduction in CHOL and SM levels with culturing time and after treatment with DAC. Panel B shows analysis of sensitive cell lipids for CHOL (a) and SM levels (b) with culturing time and following treatment with DAC. Each experiment was carried out in triplicate and data are shown as mean ± s.e.m., n = 3, *p < 0.05, between the respective untreated and treated groups in Figure 1A(a, c, e) and Figure 1B(b, d, f) between groups treated with DAC-sol vs. DAC-NGs.
While analyzing the effect of DAC treatment, we noticed that control NGs (without DAC) also seem to show marginal effect on CHOL and SM content with culturing time when compared with untreated cells (Figure 1Aa, c, e; p=N.S.), but the normalized data with respective controls (i.e., cells treated with DAC-sol vs. untreated cells and cells treated with DAC-NGs vs. cells treated with control NGs) showed a greater effect of DAC-NGs than DAC-sol in lowering SM and CHOL levels (Figure 1Ab, d). The data thus indicate reduction in CHOL-SM rafts following treatment of resistant cells with DAC (Figure 1Ab, d) and the effect of DAC-NGs is greater than with DAC-sol (Figure 1Af). Sensitive MCF-7 cells did not show any significant change in CHOL or SM content with culturing time or following treatment with DAC (Figure 1Ba, b).
Analysis of levels of phospholipids and neutral lipids following treatment with DAC
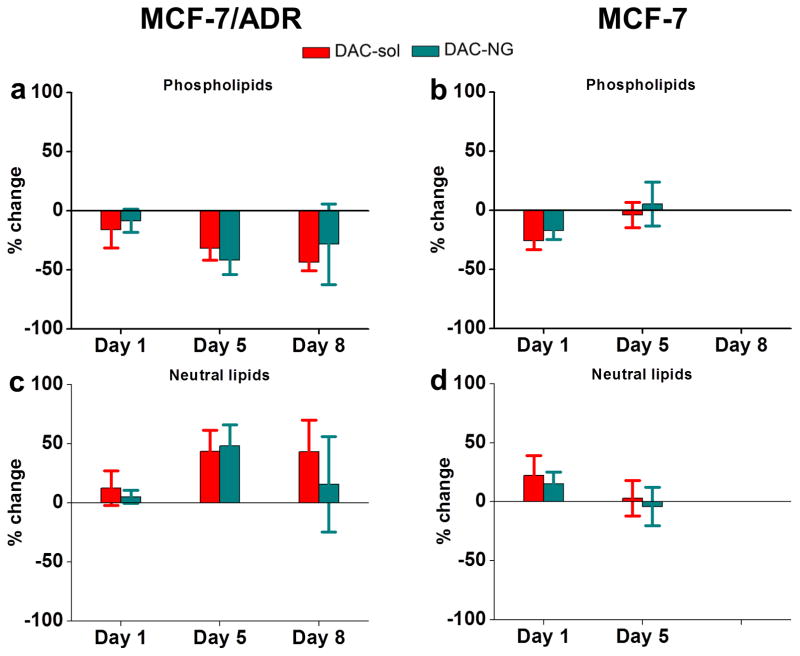
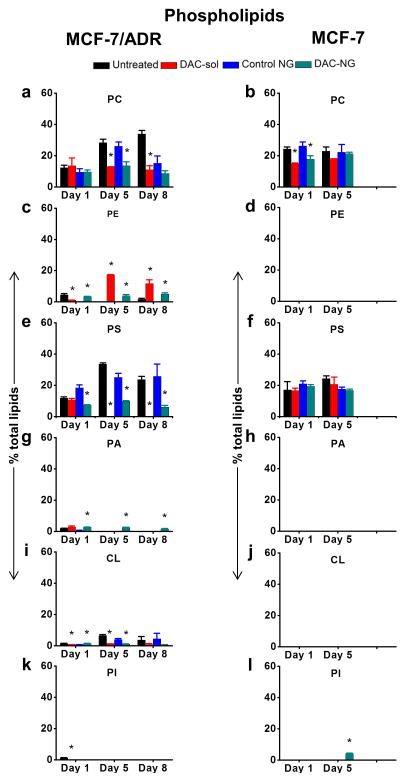
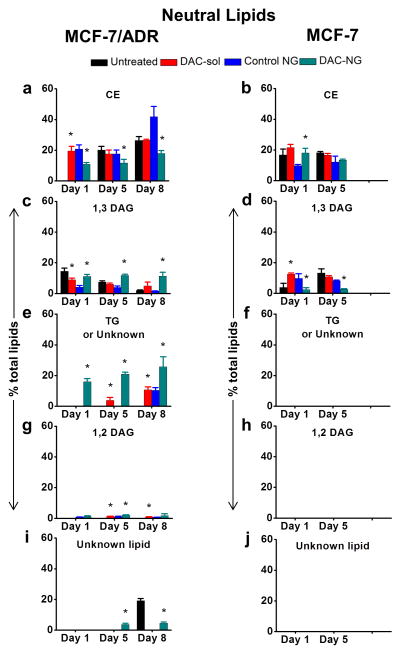
Treatment of resistant cells with DAC with the data normalized to respective controls showed a trend towards decrease in total phospholipid levels and increase in total neutral lipids but the effect of DAC treatment in sensitive cells was marginal and transient (Figure 2). Detailed lipid analysis by HPLTC showed that, most noticeably, three phospholipids, phosphatidylethanolamine (PE, Figure 3c vs. d), phosphatic acid (PA, Figure 3g vs. h), and cardiolipin (CL; Figure 3i vs. j) and three neutral lipids triglyceride (TG, could be a unknown lipid as well, Figure 4e vs. f), 1,2-diacylglycerol (1, 2-DAG; Figure 4g vs. h), and an unknown neutral lipid (Figure 4i vs. j) were present in resistant cells but not in sensitive cells.
Figure 2. Changes in total phospholipids and neutral lipids with culturing time and following treatment with DAC in resistant and sensitive cells.
Percent change in phospholipids (a, b) and neutral lipids (c, d) in resistant vs. sensitive cells following treatment with DAC-NGs or DAC-sol. Phospholipids and neutral lipids isolated at different time points post treatment were analyzed using HPTLC and data were normalized to the respective controls to calculate the percent change from the respective controls (Control NGs or culture medium). Data are mean ± s.e.m., n = 3. Data are not significantly different between DAC-NGs vs. DAC-sol.
Figure 3. Analysis of levels of individual phospholipids with culturing time and following treatment with DAC.
Cells were treated h with DAC (50 ng/mL) either as DAC-sol or DAC-NGs. Lipids were extracted at different time points and analyzed by HPTLC. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; PA, phosphatic acid; CL, cardiolipin. Data are shown as mean ± s.e.m., n = 3. *p < 0.05 between respective untreated and treated groups. Not all p-values are marked in the figure but they are as follows: Figure 3a, c and e, p-values between untreated day 1 vs. untreated day 5, untreated day 5 vs. untreated day 8 and untreated day 1 vs. untreated day 8 are significant. In Figure 3g, p-values between untreated day1 vs. untreated day 5 and untreated day 1 vs. untreated day 8 are significant. In Figure 3i, p-values between untreated day 1 vs. untreated day 5 are significant.
Figure 4. Analysis of levels of different neutral lipids with culturing time and following treatment with DAC.
Cells were treated with DAC (50 ng/mL). Lipids were extracted at different time points and analyzed by HPTLC. Data are shown as mean ± s.e.m., n = 3. *p < 0.05 between respective untreated and treated groups. DAC-sol, DAC in solution; DAC-NG, DAC in nanogel; CE, cholesterol esters; 1,2 DAG/1,3 DAG, 1,2-/1,3-diacylglycerol; TG, triglyceride(s). Not all p-values obtained between lipid levels of untreated resistant cells at different time points are not shown but are as follows: In Figure 4a, c and e, p-values between untreated day1 vs. untreated day 5, untreated day 5 vs. untreated day 8 and untreated day1 vs. untreated day 8 are significant. In Figure 4d p-values between untreated day1 vs. untreated day 5 are significant.
Treatment of resistant cells with DAC partially reversed the trend in changes in some of the individual phospholipids and neutral lipids with culturing time. The increase in the levels of phosphatidylcholine (PC, Figure 3a) and CHOL esters (CE, Figure 4a) or decrease in the levels of PA (Figure. 3g) and 1,3-diacylglycerol (1,3-DAG; Figure 4c) with culturing time in untreated resistant cells was reversed following treatment with DAC. Interestingly, certain lipids, which were not present in resistant cells prior to DAC treatment, were seen following the treatment. Particularly, prior to the treatment, TG (Figure 4e) and 1,2-DAG (Figure 4g) were not detectable in resistant cells but were seen following the treatment. Further certain lipid in resistant cells that did not change with a definitive trend with culturing time or followed a particular trend when treated with DAC. For instance, PE (Figure 3c) and CL (Figure 3i) levels increased, whereas phosphatidylserine (PS, Figure 3e) levels decreased after treatment. The most noticeable changes seen with DAC treatment were decrease in PC (Figure 3a), CEs (Figure 4a), TG (Figure 4e), 1,2-DAG (Figure 4g) and PS (Figure 3e) or increase in PA (Figure 3g), 1,3-DAG (Figure 4), PE (Figure 3c) and CL (Figure 3i) when compared with untreated cell lipids. Sensitive cells, on the other hand, did not show changes in phospholipid or neutral lipid levels or followed a certain trend with culturing time or with DAC treatment (data collected up to 5 days in MCF-7 cells) (Figure 3 and 4).
Characterization of biophysical changes in membrane lipids with culturing time and following treatment with DAC
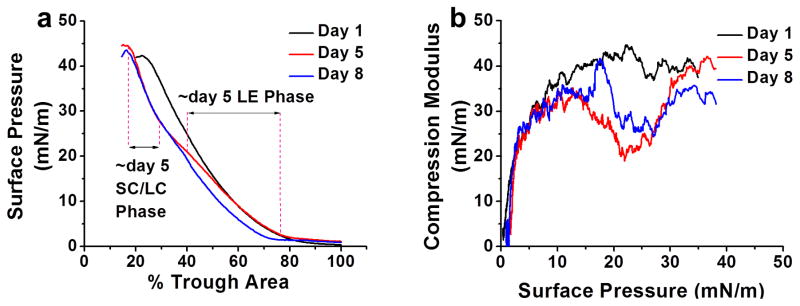
The isotherm of the resistant cell lipids showed a liquid expanded phase followed by an SC/LC phase with compression prior to collapse. The isotherm of the resistant cell lipids extracted at day 5 post culturing was seen to traverse through the isotherms of the lipids extracted on days 1 and 8 post culturing, indicative of gradual changes in the lipid profile with culturing time (Figure 5a). The A0′ (extrapolated SC/LC phase area [at zero surface pressure]) reflects that the packing density for the untreated resistant cell lipids decreases with culturing time (A0′ on days 1, 5 and 8 is at 60%, 44% and 50%, respectively) (Figure 5a). The change in compression modulus of lipids extracted at different time points shows decrease in lipid membrane fluidity at physiological SP π ~30 mN/m with culturing time.20 However, at SP π ~25 mN/m where the transition was seen, the order was day 1 less than day 5; and day 5 similar to day 8. The different parameters compiled from the isotherms of resistant cell lipids showed significant changes with culturing time, but overall data from the changes in A0′ and compression modulus indicate that both lipid packing density and membrane fluidity of resistant cell-membrane lipids increase (Figure 5a and b) with culturing time (Table 1). The isotherms of sensitive cell lipids were not characterized, since there were no significant changes in lipid composition with culturing time or following treatment with DAC. However, in our previous studies where resistant and sensitive cells were treated with DAC-sol for 24 h showed marginal changes in biophysical characteristics of membrane lipids of sensitive cells but significantly greater for resistant cells.6
Figure 5. Biophysical characterization of membrane lipids of resistant cells.
The compression isotherm (π - A) (a) and compression modulus (b) of membrane lipids extracted at different time points. The lipids were spread at different SPs, then compressed at 5 mm/min. The compression modulus was calculated from π - A isotherm data using Cs−1 = −A* (dπ/dA). The data showed an increase in lipid packing density and fluidity with culturing time.
Table 1.
Biophysical characterization of resistant cell-membrane lipids
| Parameter | Day | Untreated | DAC-sol treated | Control-NGs treated | DAC-NGs treated |
|---|---|---|---|---|---|
| Ao′ (% trough area) | 1 | 60 | 65 | 56 | 60 |
| 5 | 44 | 60 | 37 | 55 | |
| 8 | 50 | 50 | 33 | 43 | |
| Compression modulus at π30 (mN/m) | 1 | 39 | 29 | 29 | 30 |
| 5 | 36 | 36 | 25 | 27 | |
| 8 | 31 | 31 | 16 | 26 | |
| πmax (mN/m) | 1 | 42 | 39 | 45 | 42 |
| 5 | 45 | 42 | 45 | 44 | |
| 8 | 43 | 43 | 43 | 42 | |
| % trough area at πmax | 1 | 23 | 21 | 19 | 24 |
| 5 | 17 | 22 | 11 | 13 | |
| 8 | 16 | 16 | 3 | 13 |
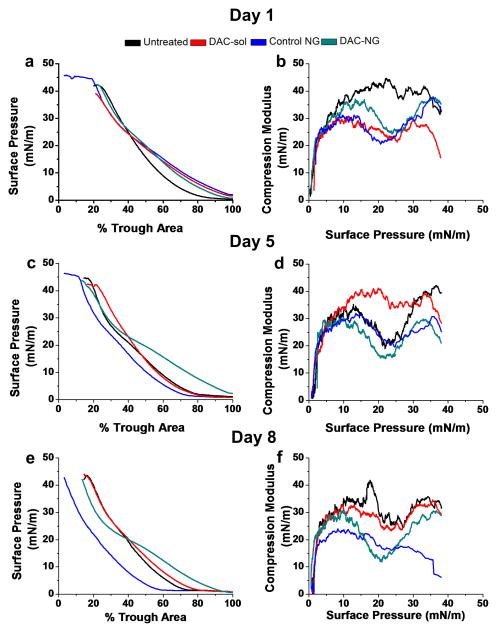
Effect of DAC treatment on the biophysical characteristics of lipids from resistant cells
The isotherms of the resistant cell lipids following treatment with DAC (both DAC-sol and DAC-NGs) showed higher A0′ (lower packing density, less compact) compared with the corresponding untreated cells (Figure 6a and c; Table 1); however, the effect of DAC-sol in increasing A0′ diminished with culturing time. The isotherms of resistant cell lipids following treatment with DAC-NGs showed only a small but significant decrease in A0′ (increase in lipid packing density on day 8 post treatment; Figure 6e; Table 1) compared with untreated cell lipids, despite the effect of control-NG itself showed a decrease in A0′ to a large extent (decrease in lipid packing density) compared with the untreated cell lipids (Figure 6e; Table 1). The most interesting finding was that resistant cell lipids treated with control NGs showed a much lower A0′ than untreated cells lipids at all culturing times (Figure 6a, c and e; Table 1). The compression modulus data (both at π = 25 and 30 mN/m) of resistant cell lipids following treatment with DAC-sol showed an increase in membrane fluidity (Figure 6b; Table 1) only on the first day of the treatment (Figure 6d and f; Table 1). Both DAC-NGs treated and control-NGs treated resistant lipids compared with corresponding untreated cell lipids (Figure 6b, d and f; Table 1) showed a sustained increase in membrane fluidity until day 8 post-treatment.
Figure 6. Effect of DAC treatment on biophysical characteristics of membrane lipids of resistant cells.
The compression isotherms (π-A) (a, c, e) and compression modulus (b, d, f) of membrane lipids extracted at different time points with DAC treatment (DAC-sol or DAC-NGs) or control (Control NGs or culture medium).
Discussion
In view of their geometry, which resembles a cone and an inverted cone, CHOL and SM together can form closely packed structures resembling a cylinder, which together form Lβ (ordered gel bilayer structures.21–22 These tightly packed Lβ structures enforce a higher structural rigidity (high lipid packing density) and lower lateral fluidity to the cell membrane.23 These Lβ structures in lipid membrane are referred to as rafts.24 In our study, resistant cell-membrane lipids showed significantly increased levels of SM with culturing time which can form CHOL-SM rafts (Figure 1A). Interestingly, lipids of sensitive cells show only marginal change in CHOL-SM levels with culturing time (Figure 1B). The point to be noted here is that CHOL and SM levels in resistant cell lipids were analyzed by HPLC whereas those in sensitive cells lipids using HPTLC, hence their relative levels in sensitive and resistant cells cannot be compared. This is because HPLC analyzes the absolute amount of a particular lipid in total lipids whereas HPTLC provides a relative amount, based on band intensity and depending upon which lipids are detectable. Therefore, in this study, it appears that CHOL and SM content is similar in both resistant and sensitive cells (Figure 1A vs. B). However, in our previous studies, where lipids in both cell lines were analyzed using the same HPTLC method, CHOL and SM levels were significantly lower in sensitive cells than in resistant cells,6 which is consistent with the literature data.25 Despite the use of different methods, the overall observation that DAC treatment has marginal effect on SM or CHOL levels of sensitive cells whereas resistant cells show significant changes remains unchanged (Figure 1A vs. B).
Since cell membranes require both high lateral fluidity and structural rigidity for their function and survival,26 the high packing density and lower fluidity in resistant cell membranes, induced by the CHOL-SM rafts, must be compensated for with other lipids that lower lipid packing density and increase fluidity to cell membranes. Hence, as culture time increased, resistant cells also showed an increase in certain phospholipids, particularly PS and PC (Figure 3), which can form Lα (liquid crystalline phase) to counter high lipid packing density and lower fluidity enforced by rafts.21–22, 27 It is known that Lα structures are relatively more fluid than Lβ structures.23 Although the geometry of PS and PC resembles a cylinder, these lipids do not form Lβ structures because of the presence of unsaturated acyl chain in the hydrophobic groups that resists formation of closely packed structures.23 It is also possible that the presence of excess CHOL that did not form rafts with SM, along with a small but significant increase in two other cone-shaped lipids, PE (Figure 3a, c) and CE (Figure 4a) can increase membrane lipid fluidity substantially as they have rigid hydrophobic tails (in CHOL and CE) or well branched tails (in PE) that do not allow the formation of closed packed structures21–23, 28 and are incompatible with Lβ structures.23 The isotherm data of resistant cell lipids with culturing time showed an increase in lipid packing density as determined from the changes in A0,′ but also an increase in membrane fluidity, which is evident from the decrease in compression modulus (Table 1). Thus the competing phenomena are happening with culturing time, i.e., increase in CHOL-SM raft structures that increase lipid packing density and hence increased membrane rigidity but at the same time there is also increased synthesis of lipids (e.g., PE, PC, CE) that increases membrane fluidity. Despite increased membrane fluidity with culturing time, it is most likely that resistant cells become more resistant as they divide due to increasing levels of CHOL-SM rafts, which can markedly influence drug transport.29
The most conspicuous effect of DAC treatment on resistant cells was a simultaneous decrease in CHOL and SM content with culturing time (Figure 1Aa–d), and hence resulting in decrease in the levels of CHOL-SM rafts (Figure 1Ae–f). The effect of DAC-NGs sol in lowering levels of CHOL-SM rafts was greater than DAC-sol which could be because of the instability of DAC in culture medium (half-life =~17 h) (Figure 1Ab) 30 whereas DAC-NGs stabilizing and facilitating DAC delivery is more effective (Figure 1Ae, f). In our previous studies, we have demonstrated that DAC-NGs is more effective in inhibiting proliferation of MCF-7/ADR and other cancer cells (resistant leukemia and melanoma cells), whereas the effect of DAC-sol was transient, suggesting that NG formulation protects the enclosed load of DAC, thereby enhancing its effect.17
Although control-NGs showed marginal effect on lipid levels when compared with untreated cells, it had no effect on the levels of CHOL-SM rafts (Figure 1A). We speculate that unusually higher lipid packing density and also fluidity in the control NG-treated cell lipids compared with the untreated cell lipids (Figure 6) is probably a consequence of some polymeric chain segments of NGs or NG fragments following their disintegration are adsorbed on the lipids. Another explanation may be that these segments could interfere with the lipid packing density by altering electrostatic repulsion between the head groups of the lipids. However, DAC-NGs apparently seems to be less effective in increasing lipid fluidity (compression modulus of DAC-NG vs. control-NG= 26 vs. 16 mN/m respectively on day 8 post treatment; Table 1). Considering that there is no significant difference in physical characteristics (size and zeta potential) of control NGs and DAC-NGs,17 it is hard to explain the unusual effect of control NGs. However, it is possible that cellular interactions and uptake of control NGs and DAC-NGs could be different.
Control NGs might mainly remain adhered to the cell membrane because of their bioadhesive property whereas DAC-NGs might internalize due to the effect of DAC (Figure 1A). As discussed earlier, resistant cell membrane lipids are more resistant to endocytic uptake and treatment with DAC can regain their endocytic function due to changes in the lipid profile.6 Since the treatment of resistant cells with DAC-NGs alters CHOL-SM rafts and other lipids suggesting their intracellular uptake to influence lipid biosynthesis. Control NGs on the other hand may remain with the lipids even after going through the extraction procedure, affecting lipid-lipid interactions and hence biophysical properties of lipid membrane properties. Further studies looking at cellular uptake of control NGs and DAC-NGs in resistant cells could explain the effect of control NGs and DAC-NGs on biophysical properties of membrane lipids.
The overall effect of DAC treatment, based on the isotherm and lipid analysis data, was a decrease in lipid packing density (Figure 6a, c and d) due to lowering levels of rafts and an increase in membrane fluidity (6 b, d and f) due to decrease in lipids that form Lα structures (PC, PS and CE)23 and increase in lipids (PE and PA) that form nonlamellar structures. The nonlamellar structures of PE and PA can promote endocytosis in resistant cells, the primary mechanism of uptake of NP-based drug delivery systems.5 Further, since treatment with DAC-NGs lowers SM levels more than CHOL levels (Figure 1e and f), the relative concentration of CHOL will be higher in resistant cells following DAC-NG treatment, which by itself can disrupt the raft structures because of the incompatibility of CHOL (a cone-shaped lipid) and Lβ raft structures.31
A higher reduction in levels SM than of CHOL following DAC-NG treatment of resistant cells would be expected to decrease lipid packing density and increase fluidity, and this assumption was corroborated from our isotherm data (Table 1). A particularly noticeable effect of altered lipid levels in resistant cells induced by DAC treatment was (a) an increase in the total levels of phospholipids, which have been reported to play a major role in drug transport by modulating drug diffusion across the cell membrane,32 as well as in sorting mechanisms in endocytosis33 and (b) decreases in levels of CHOL-SM rafts, which can lead to increased drug permeation,5–6 lowered P-glycoprotein activity associated with drug efflux,34 and increased ceramide formation on the cell membrane,35 all of which are key factors in reversing drug resistance in cancer cells.13
Our data indicate there is a significant difference in the composition of the membrane lipids of sensitive vs. resistant cells (Figures 1–3). It is interesting to note that several lipids, not present prior to DAC treatment, do appear following treatment. Thus, it may be that genes for SMase, which are known to be silenced in resistant cells,16 are not the only genes affected; genes for several other lipids (e.g., PE, PA, CL, phosphatidylinositol, and 1,2-DAG) may be silenced as well (Figures 3–4). Most noticeable are the lipids responsible for facilitating drug transport and endocytic function,6 suggesting that the adoptive epigenetic mechanism of cancer cells is to protect themselves from the cytotoxic effects of anticancer drugs by altering lipid synthesis to prevent internalization such drugs as the cells develop drug resistance.
Densely packed lipids are resistant to the bending required for budding that is essential for the formation of curved membranes during endocytosis,36 the primary mechanism of uptake of nanoparticle-based drug delivery systems.5 Increase in the concentration of non-lamellar lipids like PE/PA lipids can facilitate budding, thereby increasing endocytosis following treatment of resistant cells with epigenetic drugs.37 Specifically, decrease in SM and an increase in PE density facilitate the biomechanical torque needed to generate membrane budding, thereby increasing endocytosis.37 In our recent study, we determined biomechanics and thermodynamics of nanoparticle interactions of different surface properties with resistant and sensitive cell membrane lipids and how that translates to cellular uptake of nanoparticles. Our data demonstrated that it is not only the surface of nanoparticles but also membrane fluidity plays a significant role in cellular uptake of nanoparticles and their subsequent escape from endosomes. Confocal imaging data using membrane and endosomal markers demonstrated significantly greater uptake and escape of nanoparticles in sensitive cells than in resistant cells, as membrane rigidity of resistant cells affected the uptake and subsequent escape of nanoparticles from endosomes.38
Multidrug resistance is a multifactorial phenomenon. The major known mechanisms of drug resistance include, reduced drug influx, increased drug efflux via permeability glycoprotein (P- P-gp),39–40 drug entrapment in intracellular vesicles,41–42 and altered or defective apoptotic pathways. Most of these mechanisms are associated with change in membrane lipids composition. The reduced drug influx has been shown to be related to phospholipid levels and also CHOL-SM rafts. In our work, a particularly noticeable effect of altered lipid levels in resistant cells induced by DAC treatment was an increase in the total levels of certain phospholipids and neutral lipids, which have been reported to play a major role in drug transport by modulating drug diffusion across the cell membrane,32 as well as in sorting mechanisms in endocytosis.33 The decrease in CHOL-SM rafts (Lβ structures) following DAC-NG treatment is expected to allow better permeation of drugs across lipid membranes, which otherwise might get entrapped due to hydrophobic interactions and drug partitioning in membrane.
The P-gp activity is shown to be dependent on the cholesterol and SM levels.43–45 P-gp was reported to be more active when localized in lipid rafts rich in SM and CHOL than outside the rafts.46 We have recently reported that DAC-treated resistant cells show reduced level of P-gp than in entreated resistant cells, thus signifying the lipid membrane composition on P-gp activity. 6 Based on our data from this work, we infer that the sustained depiction of CHOL-SM rafts following DAC-treatment to resistant cells also results in depletion or reduced in P-gp activity.6
The drug sequestration in intracellular acidic compartments, such as lysosomes, recycling endosomes and the trans-Golgi network has been known.47–49 However, recent reports show that the negatively charged lipids like PS present in the inner leaflet show diminished head group charge at acidic pH.50–51 The diminished head group charge leads to formation of close packed structures due to decreased electrostatic repulsion between head groups, which hinder drug diffusion from acidic intracellular compartments.13, 50–51 In our work, DAC-NG treatment to resistant cells decreases negatively charged lipid- PS level in a sustained manner but increases the lipid levels of PA (also negatively charged). However quantitatively, decrease in PS levels is more than increase in PA levels. Thus it is conceivable that the decrease in PS levels will help diffusion of drugs from acidic compartments.
Thus there could be multiple mechanisms via which changes in lipid composition following treatment with epigenetic drugs can overcome drug resistance. Hence, it would be of further interest to determine how effective DAC-NG would be in overcoming drug resistance if administered in combination with other anticancer drugs by regaining drug transport or endocytic function. NGs can potentially protect DAC from rapid metabolism and clearance thus enhancing its efficacy to treat different cancers either alone or in combination therapy.
Conclusions
Overall, our results demonstrate that (a) the lipid profile of resistant cells (but not sensitive cells) changes with culturing time; (b) the effect of DAC treatment on changes in lipid profile is more specific in resistant cells than in sensitive cells, indicating that epigenetic changes are responsible for altered lipid synthesis in resistant cells; and (c) the effect of DAC is greater with DAC-NGs than with DAC-sol. The increased fluidity of membrane lipids following DAC treatment may facilitate drug transport and that could be a key means of overcoming drug resistance.
Acknowledgments
This study is funded by grant R01 CA149359 (to VL) from the National Cancer Institute of the National Institutes of Health.
ABBREVIATIONS
- CHOL
Cholesterol
- CL
Cardiolipin
- DAC
5-aza-2′-deoxycytidine (decitabine)
- DAC-NG
DAC in nanogel formulation
- DAC-sol
DAC in solution
- 1,2-DAG
1,2-diacylglycerol
- 1,3-DAG
1,3-diacylglycerol (1,3-DAG
- EDTA
Ethylenediaminetetraacetic acid
- HPLC
High-performance liquid chromatography
- HPTLC
High-performance thin-layer chromatography
- PA
Phosphatic acid
- PC
Phosphatidylcholine
- PE
Phosphatidylethanolamine
- PS
Phosphatidylserine
- SC/LC
Solid condensed/liquid condensed
- SM
Sphingomyelin
- SMase
Sphingomyelinase
- SP
Surface pressure
- TG
Triglycerides
- Tris
Tris(hydroxymethyl)aminomethane
References
- 1.Wymann MP, Schneiter R. Lipid Signalling in Disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 2.Grösch S, Schiffmann S, Geisslinger G. Chain Length-Specific Properties of Ceramides. Prog Lipid Res. 2012;51:50–62. doi: 10.1016/j.plipres.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Clay AT, Sharom FJ. Lipid Bilayer Properties Control Membrane Partitioning, Binding, and Transport of P-Glycoprotein Substrates. Biochemistry. 2013;52:343–354. doi: 10.1021/bi301532c. [DOI] [PubMed] [Google Scholar]
- 4.Hannun YA, Obeid LM. The Ceramide-Centric Universe of Lipid-Mediated Cell Regulation: Stress Encounters of the Lipid Kind. J Biol Chem. 2002;277:25847–25850. doi: 10.1074/jbc.R200008200. [DOI] [PubMed] [Google Scholar]
- 5.Peetla C, Bhave R, Vijayaraghavalu S, Stine A, Kooijman E, Labhasetwar V. Drug Resistance in Breast Cancer Cells: Biophysical Characterization of and Doxorubicin Interactions with Membrane Lipids. Mol Pharmaceutics. 2010;7:2334–2348. doi: 10.1021/mp100308n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijayaraghavalu S, Peetla C, Lu S, Labhasetwar V. Epigenetic Modulation of the Biophysical Properties of Drug-Resistant Cell Lipids to Restore Drug Transport and Endocytic Functions. Mol Pharmaceutics. 2012;9:2730–2742. doi: 10.1021/mp300281t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewers H, Helenius A. Lipid-Mediated Endocytosis. Cold Spring Harbor Perspect Biol. 2011;3:a004721. doi: 10.1101/cshperspect.a004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramu A, Glaubiger D, Magrath IT, Joshi A. Plasma Membrane Lipid Structural Order in Doxorubicin-Sensitive and -Resistant P388 Cells. Cancer Res. 1983;43:5533–5537. [PubMed] [Google Scholar]
- 9.Ramu A, Glaubiger D, Weintraub H. Differences in Lipid Composition of Doxorubicin-Sensitive and -Resistant P388 Cells. Cancer Treat Rep. 1984;68:637–641. [PubMed] [Google Scholar]
- 10.May GL, Wright LC, Dyne M, MacKinnon WB, Fox RM, Mountford CE. Plasma-Membrane Lipid-Composition of Vinblastine Sensitive and Resistant Human-Leukaemic Lymphoblasts. Int J Cancer. 1988;42:728–733. doi: 10.1002/ijc.2910420517. [DOI] [PubMed] [Google Scholar]
- 11.Gouazé V, Mirault ME, Carpentier S, Salvayre R, Levade T, Andrieu-Abadie N. Glutathione Peroxidase-1 Overexpression Prevents Ceramide Production and Partially Inhibits Apoptosis in Doxorubicin-Treated Human Breast Carcinoma Cells. Mol Pharmacol. 2001;60:488–496. [PubMed] [Google Scholar]
- 12.Rauch C. Toward a Mechanical Control of Drug Delivery. On the Relationship between Lipinski’s 2nd Rule and Cytosolic Ph Changes in Doxorubicin Resistance Levels in Cancer Cells: A Comparison to Published Data. Eur Biophys J. 2009;38:829–846. doi: 10.1007/s00249-009-0429-x. [DOI] [PubMed] [Google Scholar]
- 13.Peetla C, Vijayaraghavalu S, Labhasetwar V. Biophysics of Cell Membrane Lipids in Cancer Drug Resistance: Implications for Drug Transport and Drug Delivery with Nanoparticles. Adv Drug Deliv Rev. 2013;65:1686–1698. doi: 10.1016/j.addr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hendrich AB, Michalak K. Lipids as a Target for Drugs Modulating Multidrug Resistance of Cancer Cells. Curr Drug Targets. 2003;4:23–30. doi: 10.2174/1389450033347172. [DOI] [PubMed] [Google Scholar]
- 15.Pallarés-Trujillo J, López-Soriano FJ, Argilés JM. Lipids: A Key Role in Multidrug Resistance? Int J Oncol. 2000;16:783–798. doi: 10.3892/ijo.16.4.783. [DOI] [PubMed] [Google Scholar]
- 16.Demircan B, Dyer LM, Gerace M, Lobenhofer EK, Robertson KD, Brown KD. Comparative Epigenomics of Human and Mouse Mammary Tumors. Genes Chromosome Cancer. 2009;48:83–97. doi: 10.1002/gcc.20620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vijayaraghavalu S, Labhasetwar V. Efficacy of Decitabine-Loaded Nanogels in Overcoming Cancer Drug Resistance Is Mediated Via Sustained DNA Methyltransferase 1 (Dnmt1) Depletion. Cancer Lett. 2013;331:122–129. doi: 10.1016/j.canlet.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdick D, Soreghan B, Kwon M, Kosmoski J, Knauer M, Henschen A, Yates J, Cotman C, Glabe C. Assembly and Aggregation Properties of Synthetic Alzheimers A4/Beta Amyloid Peptide Analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 19.Narváez-Rivas M, Gallardo E, Ríos JJ, León-Camacho M. A New High-Performance Liquid Chromatographic Method with Evaporative Light Scattering Detector for the Analysis of Phospholipids. Application to Iberian Pig Subcutaneous Fat. J Chromatogr. 2011;1218:3453–3458. doi: 10.1016/j.chroma.2011.03.067. [DOI] [PubMed] [Google Scholar]
- 20.Zwaal RFA, Demel RA, Roelofsen B, van Deenen LLM. The Lipid Bilayer Concept of Cell Membranes. Trends Biochem Sci. 1976;1:112–114. [Google Scholar]
- 21.Cullis PR, de Kruijff B. Lipid Polymorphism and the Functional Roles of Lipids in Biological Membranes. Biochimica Et Biophysica Acta. 1979;559:399–420. doi: 10.1016/0304-4157(79)90012-1. [DOI] [PubMed] [Google Scholar]
- 22.Frolov VA, Shnyrova AV, Zimmerberg J. Lipid Polymorphisms and Membrane Shape. Cold Spring Harbor Perspect Biol. 2011;3:a004747. doi: 10.1101/cshperspect.a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins and Membranes. 4. Elsevier Science B.V; Amsterdam: 2002. [Google Scholar]
- 24.George KS, Wu S. Lipid Raft: A Floating Island of Death or Survival. Toxicol Appl Pharmacol. 2012;259:311–9. doi: 10.1016/j.taap.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Todor IN, Lukyanova NY, Chekhun VF. The Lipid Content of Cisplatin- and Doxorubicin-Resistant Mcf-7 Human Breast Cancer Cells. Exp Oncol. 2012;34:97–100. [PubMed] [Google Scholar]
- 26.Espinosa G, López-Montero I, Monroy F, Langevin D. Shear Rheology of Lipid Monolayers and Insights on Membrane Fluidity. Proc Natl Acad Sci USA. 2011;108:6008–6013. doi: 10.1073/pnas.1018572108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cullis PR, Hope MJ, Tilcock CPS. Lipid Polymorphism and the Roles of Lipids in Membranes. Chem Phys Lipids. 1986;40:127–144. doi: 10.1016/0009-3084(86)90067-8. [DOI] [PubMed] [Google Scholar]
- 28.Chandrasekar K, Vijay R, Baskar G. Ionic Polymeric Amphiphiles with Cholesterol Mesogen: Adsorption and Organization Characteristics at the Air/Water Interface from Langmuir Film Balance Studies. Biomacromolecules. 2008;9:1264–1272. doi: 10.1021/bm701252y. [DOI] [PubMed] [Google Scholar]
- 29.Lavie Y, Liscovitch M. Changes in Lipid and Protein Constituents of Rafts and Caveolae in Multidrug Resistant Cancer Cells and Their Functional Consequences. Glycoconj J. 2000;17:253–259. doi: 10.1023/a:1026553626537. [DOI] [PubMed] [Google Scholar]
- 30.Covey JM, Zaharko DS. Effects of Dose and Duration of Exposure on 5-Aza-2′-Deoxycytidine Cytotoxicity for L1210 Leukemia in Vitro. Cancer Treat Rep. 1984;68:1475–1481. [PubMed] [Google Scholar]
- 31.Crane JM, Tamm LK. Role of Cholesterol in the Formation and Nature of Lipid Rafts in Planar and Spherical Model Membranes. Biophys J. 2004;86:2965–2979. doi: 10.1016/S0006-3495(04)74347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown R, Plumb JA. Demethylation of DNA by Decitabine in Cancer Chemotherapy. Expert Rev Anticancer Ther. 2004;4:501–510. doi: 10.1586/14737140.4.4.501. [DOI] [PubMed] [Google Scholar]
- 33.Gros C, Fahy J, Halby L, Dufau I, Erdmann A, Gregoire JM, Ausseil F, Vispe S, Arimondo PB. DNA Methylation Inhibitors in Cancer: Recent and Future Approaches. Biochimie. 2012;94:2280–2296. doi: 10.1016/j.biochi.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 34.Flotho C, Claus R, Batz C, Schneider M, Sandrock I, Ihde S, Plass C, Niemeyer CM, Lübbert M. The DNA Methyltransferase Inhibitors Azacitidine, Decitabine and Zebularine Exert Differential Effects on Cancer Gene Expression in Acute Myeloid Leukemia Cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- 35.Kilkus J, Goswami R, Testai FD, Dawson G. Ceramide in Rafts (Detergent-Insoluble Fraction) Mediates Cell Death in Neurotumor Cell Lines. J Neurosci Res. 2003;72:65–75. doi: 10.1002/jnr.10549. [DOI] [PubMed] [Google Scholar]
- 36.Maxfield FR, McGraw TE. Endocytic Recycling. Nat Rev Mol Cell Biol. 2004;5:121– 132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 37.Rauch C, Farge E. Endocytosis Switch Controlled by Transmembrane Osmotic Pressure and Phospholipid Number Asymmetry. Biophys J. 2000;78:3036–3047. doi: 10.1016/S0006-3495(00)76842-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peetla C, Jin S, Weimer J, Elegbede A, Labhasetwar V. Biomechanics and Thermodynamics of Nanoparticle Interactions with Plasma and Endosomal Membrane Lipids in Cellular Uptake and Endosomal Escape. Langmuir. 2014;30:7522–7532. doi: 10.1021/la5015219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting Multidrug Resistance in Cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 40.Perez-Tomas R. Multidrug Resistance: Retrospect and Prospects in Anti-Cancer Drug Treatment. Curr Med Chem. 2006;13:1859–1876. doi: 10.2174/092986706777585077. [DOI] [PubMed] [Google Scholar]
- 41.Larsen AK, Skladanowski A. Cellular Resistance to Topoisomerase-Targeted Drugs: From Drug Uptake to Cell Death. Biochim Biophys Acta. 1998;1400:257–274. doi: 10.1016/s0167-4781(98)00140-7. [DOI] [PubMed] [Google Scholar]
- 42.Gottesman MM. Mechanisms of Cancer Drug Resistance. Annu Rev Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 43.Troost J, Lindenmaier H, Haefeli WE, Weiss J. Modulation of Cellular Cholesterol Alters P-Glycoprotein Activity in Multidrug-Resistant Cells. Mol Pharmacol. 2004;66:1332–13339. doi: 10.1124/mol.104.002329. [DOI] [PubMed] [Google Scholar]
- 44.Gayet L, Dayan G, Barakat S, Labialle S, Michaud M, Cogne S, Mazane A, Coleman AW, Rigal D, Baggetto LG. Control of P-Glycoprotein Activity by Membrane Cholesterol Amounts and Their Relation to Multidrug Resistance in Human Cem Leukemia Cells. Biochemistry. 2005;44:4499–4509. doi: 10.1021/bi048669w. [DOI] [PubMed] [Google Scholar]
- 45.Kopecka J, Campia I, Olivero P, Pescarmona G, Ghigo D, Bosia A, Riganti C. A Ldl-Masked Liposomal-Doxorubicin Reverses Drug Resistance in Human Cancer Cells. J Control Release. 2011;149:196–205. doi: 10.1016/j.jconrel.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 46.Ghetie MA, Marches R, Kufert S, Vitetta ES. Blood. 2004;104:178. doi: 10.1182/blood-2003-12-4255. [DOI] [PubMed] [Google Scholar]
- 47.Hindenburg AA, Baker MA, Gleyzer E, Stewart VJ, Case N, Taub RN. Effect of Verapamil and Other Agents on the Distribution of Anthracyclines and on Reversal of Drug-Resistance. Cancer Res. 1987;47:1421–1425. [PubMed] [Google Scholar]
- 48.Meschini S, Molinari A, Calcabrini A, Citro G, Arancia G. Intracellular-Localization of the Antitumor Drug Adriamycin in Living Cultured-Cells - a Confocal Microscopy Study. J Microsc. 1994;176:204–210. doi: 10.1111/j.1365-2818.1994.tb03516.x. [DOI] [PubMed] [Google Scholar]
- 49.Simon SM, Schindler M. Cell Biological Mechanisms of Multidrug-Resistance in Tumors. Proc Natl Acad Sci US A. 1994;91:3497–3504. doi: 10.1073/pnas.91.9.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petelska AD, Figaszewski ZA. Effect of Ph on the Interfacial Tension of Bilayer Lipid Membrane Formed from Phosphatidylcholine or Phosphatidylserine. Biochim Biophys Acta. 2002;1561:135–146. doi: 10.1016/s0005-2736(01)00463-1. [DOI] [PubMed] [Google Scholar]
- 51.Petelska AD, Figaszewski ZA. Effect of Ph on the Interfacial Tension of Lipid Bilayer Membrane. Biophys J. 2000;78:812–817. doi: 10.1016/S0006-3495(00)76638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]