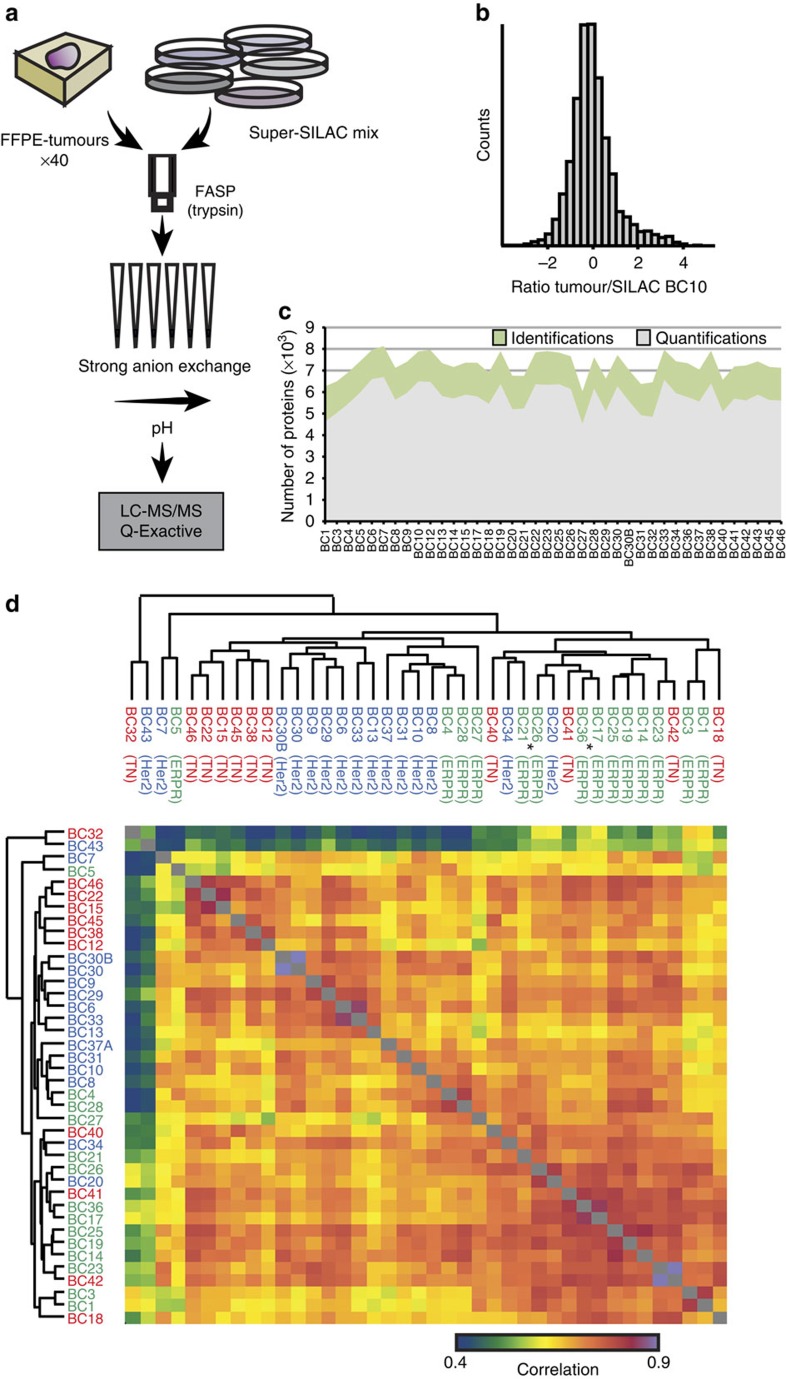
Figure 1. Super-SILAC-based quantitative proteomics of breast cancer clinical samples.
(a) Proteomics workflow involved combination of the super-SILAC mix with FFPE tumour samples, followed by FASP digestion with trypsin, peptide fractionation and analysis on the Q Exactive MS. (b) Ratio distribution between the tumour proteome (of one representative tumour) and the super-SILAC mix showed overall narrow distribution that enables accurate ratio determination. (c) Plot shows the number of identified and quantified proteins in each tumour sample. (d) Hierarchical clustering of Pearson correlations of breast cancer samples shows high diversity between tumour samples, with only partial co-clustering of samples of the same classical subtype. Two triple-positive tumours are marked with *.