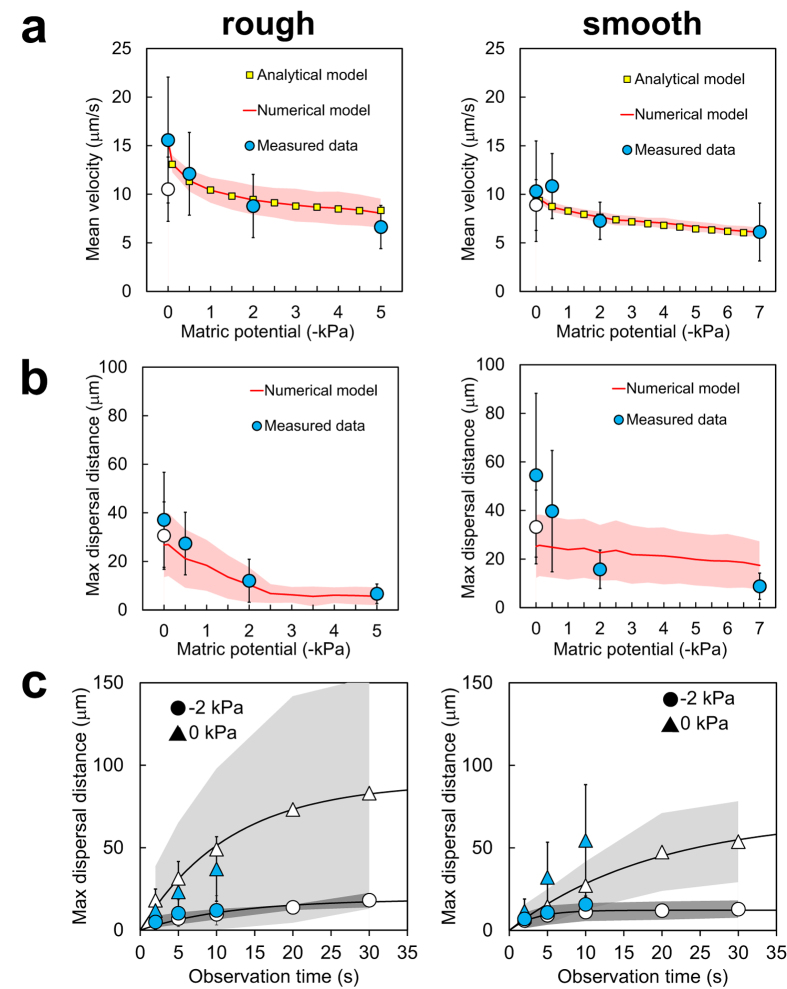
Figure 5. Quantification of Pseudomonas protegens flagellar motility on PSM and comparison with model simulations.
We tested PSM with either ‘rough’ (left panels) or ‘smooth’ (righ panels) surface characteristics. (a) Mean swimming velocity, which is the average of calculated swimming speeds >3 μm/s (the method detection limit). (b,c) Maximal dispersal distance, which is the radius of a circle centered at the initial cell position at time 0 and reaching to the most distant position occupied by the cell during the observation time (see Fig. 4a). (a,b) Circles show average values calculated from n independent swimming trajectories (observation time is 10 s): Rough: n = 20 (0 kPa), n = 20 (–0.5 kPa), n = 17 (–2 kPa), n = 15 (–5 kPa), n = 21 (back to 0 kPa); Smooth: n = 19 (0 kPa), n = 19 (–0.5 kPa), n = 17 (–2 kPa), n = 20 (–7 kPa), n = 24 (back to 0 kPa). Error bars are 1 standard deviation (SD). Matric potential was sequentially lowered (0 kPa, –0.5 kPa, etc.) and finally brought back to 0 kPa (indicated by an open circle). We used an analytical model for flagellar motility that takes into account the hydration-dependent resistive forces acting on a cell (see Methods). With numerical simulations, shaded area is ±1 SD calculated from 100 realizations. (c) Effects of observation time on the measured maximal dispersal distance at 0 kPa and –2 kPa. Blue (closed) symbols show 10 s data used in (b) and additional values for 2 s and 5 s observations. Error bars are ±1 SD. Open symbols show results from replicate experiments with observation time extended to 30 s. Rough: n = 23 (0 kPa), n = 4 (–2 kPa); smooth: n = 16 (0 kPa), n = 12 (–2 kPa). Shaded areas are ±1 SD.