Table 1. List of parameter values.
| Parameter | Definition | Value | Ref. |
|---|---|---|---|
| a1 | Scaling factor between NADH consumption and change in membrane voltage | 20 | This work |
| a2 | Scaling factor between ATP production by ATPase and change in membrane voltage | 3.43 | 28 |
| α | Volumic ratio between the endoplasmic reticulum and the cytosol | 0.1 | 40 |
| αc | Factor taking cytosolic ADP and ATP buffering into account | 0.111 | 27 |
| αm | Factor taking mitochondrial ADP and ATP buffering into account | 0.139 | 27 |
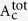 |
Total concentration in cytosolic adenine nucleotides | 2500 μM | 73 |
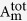 |
Total concentration in mitochondrial adenine nucleotides | 15000 μM | 28 |
| b | Ca2+ leak from the endoplasmic reticulum | 0.01 | This work |
| Cp | Mitochondrial inner membrane capacitance divided by F | 1.8 μM.mV−1 | 28 |
| δ | Volumic ratio between the mitochondria and the cytosol | 0.0733 | 28 |
| F | Faraday constant | 96480 C.mol−1 | |
| fc | Fraction of free over buffer-bound Ca2+ in the cytosol | 0.01 | 27 |
| fER | Fraction of free over buffer-bound Ca2+ in the ER | 0.01 | 27 |
| fm | Fraction of free over buffer-bound Ca2+ in mitochondria | 0.0003 | 27 |
| k1 | Rate constant of the Ca2+ flux through IP3R | 30 s−1 | This work |
| K1 | Dissociation constant for Ca2+ translocation by the MCU | 6 μM | This work |
| K2 | Dissociation constant for MCU activation by Ca2+ | 0.38 μM | 20 |
| Ka | Dissociation constant of Ca2+ from the activating site of the IP3R | 0.3 μM | 74 |
| KAGC | Dissociation constant of Ca2+ from AGC | 0.14 μM | 50 |
| Ke | Dissociation constant of ATP from SERCA pumps | 0.05 μM | 41 |
| kGLY | Velocity of glycolysis (empirical) | 450 μM.s−1 | 28 |
| Kh | Michaelis-Menten constant for ATP hydrolysis | 1000 μM | This work |
| kHYD | Maximal rate of ATP hydrolysis | 100 μM.s−1 | This work |
| Ki | Dissociation constant of IP3 binding from its receptor | 1 μM | 54 |
| k− | Rate constant of Ca2+ dissociation from the inactivating site of the IP3 receptor | 0.02 s−1 | 74 |
| ko | Rate constant of NADH oxidation by ETC | 600 μM.s−1 | 28 |
| Kp | Dissociation constant of Ca2+ from SERCA | 0.35 μM | 54 |
| k+ | Rate constant of Ca2+ binding to the inhibiting site of the IP3R | 20 μM−4.s−1 | This work |
| kx | Rate constant of bidirectional Ca2+ leak from mitochondria | 0.008 s−1 | This work |
| L | Allosteric equilibrium constant for uniporter conformations | 50 | 21 |
| na | Hill coefficient of Ca2+ binding to the activating site of the IP3R | 3 | 74 |
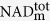 |
Total concentration of mitochondrial pyridine nucleotides | 250 μM | This work |
| ni | Hill coefficient of Ca2+ binding to the inhibiting site of the IP3R | 4 | 74 |
| p1 | Voltage dependence coefficient of MCU activity | 0.1 mV−1 | This work |
| p2 | Voltage dependence coefficient of NCX activity | 0.016 mV−1 | This work |
| p3 | Voltage dependence coefficient of calcium leak | 0.05 mV−1 | This work |
| p4 | Voltage dependence coefficient of AGC activity | 0.01 mV−1 | This work |
| q1 | Michaelis-Menten-like constant for NAD+ consumption by the Krebs cycle | 1 | 28 |
| q2 | S0.5 value for activation the Krebs cycle by Ca2+ | 0.1 μM | This work |
| S0.5 value for indirect inhibition of the AGC by cytosolic Ca2+ | 0.1 μM | This work | |
| q3 | Michaelis-Menten constant for NADH consumption by the ETC | 100 μM | 28 |
| q4 | Voltage dependence coefficient 1 of ETC activity | 177 mV | 28 |
| q5 | Voltage dependence coefficient 2 of ETC activity | 5 mV | 28 |
| q6 | Inhibition constant of ATPase activity by ATP | 10000 μM | 28 |
| q7 | Voltage dependence coefficient of ATPase activity | 190 mV | 28 |
| q8 | Voltage dependence coefficient of ATPase activity | 8.5 mV | 28 |
| q9 | Voltage dependence of the proton leak | 2 μM.s−1.mV−1 | 28 |
| q10 | Rate constant of the voltage-independent proton leak | −30 μM.s−1 | 28 |
| R | Perfect gas constant | 8315 mJ.mol−1.K−1 | |
| T | Temperature | 310.16 K | |
| VANT | Rate constant of the adenine nucleotide translocator | 5000 μM.s−1 | 28 |
| VAGC | Rate constant of NADH production via malate-aspartate shuttle | 25 μM.s−1 | This work |
| VF1FO | Rate constant of the F1FO ATPase | 35000 μM.s−1 | 28 |
| VMCU | Rate constant of the MCU | 0.0006 μM.s−1 | This work |
| VNCX | Rate constant of the NCX | 0.35 μM.s−1 | This work |
| Vp | Rate constant of the SERCA pumps | 120 μM.s−1 | 54 |
Fluxes are defined with respect to the volumes of the cytoplasm, the ER or the mitochondria as indicated in the text. Thus, as described in Fall and Keizer27, the values of the rate constants obtained experimentally and expressed in nmol.(mg.s)−1 are multiplied by the total protein amounts for each compartment and divided by the volume of the compartment.
