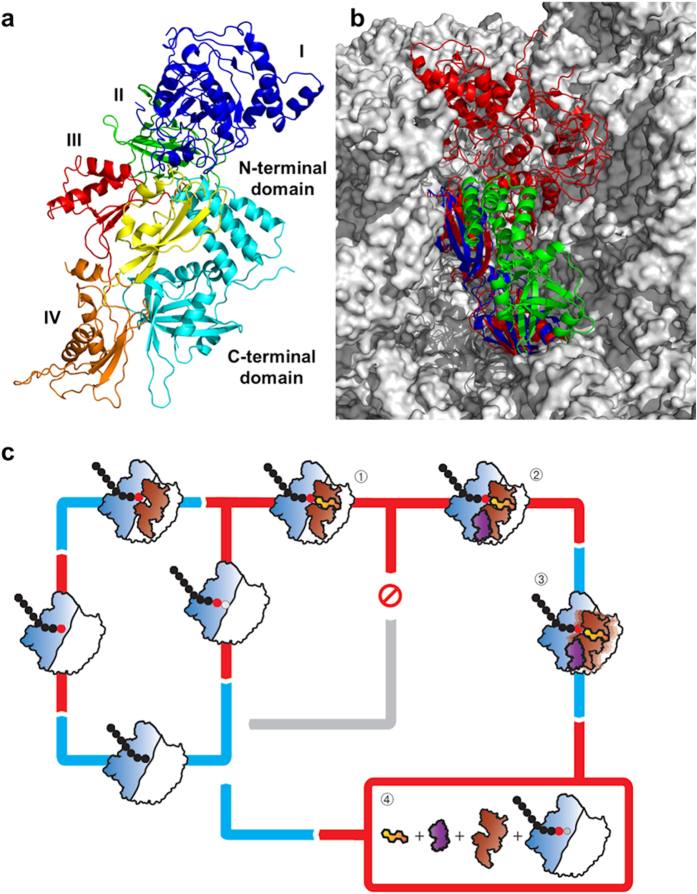
Figure 6. Structure of the FusB•EF-G complex, and the mechanism of FA resistance.
(a) A model of FusB binding to full length EF-G produced by aligning the FusB•EF-GC3 structural model with the crystal structure of ribosome bound EF-G. EF-G is colored by domain and FusB is shown in cyan. (b) The model of the FusB•EF-GC3 complex docked (by sequence alignment of domains IV and V) onto EF-G bound to the ribosome in the post-translocational state. EF-GC3 is shown in blue, FusB in green, EF-G bound to the ribosome in red, and the ribosome in grey. (c) Suggested mechanism of FusB-mediated resistance to FA. EF-G (brown) binds to the ribosome (blue and white) after peptide bond formation between the A and P site amino acids and mediates translocation to the P and E sites. EF-G then dissociates making the A site available for binding of the next tRNA. In the presence of the drug, FA (yellow) binds to EF-G on the ribosome in the post-translocation state (1) and stalls protein synthesis by preventing EF-G release. Binding of FusB (purple) to EF-G in stalled complexes (2) induces a conformational change in domains IV and V of EF-G and a change in the dynamics of domain III (3). Either this conformational change is sufficient to promote release of EF-G or the dynamics within domain III allow the C-terminal domains of EF-G to more readily adopt the conformation relative to domains I and II required for release, without the requirement for transmission of conformational change from domains I and II. This results in dissociation of EF-G from the ribosome, thereby allowing protein synthesis to continue (4).