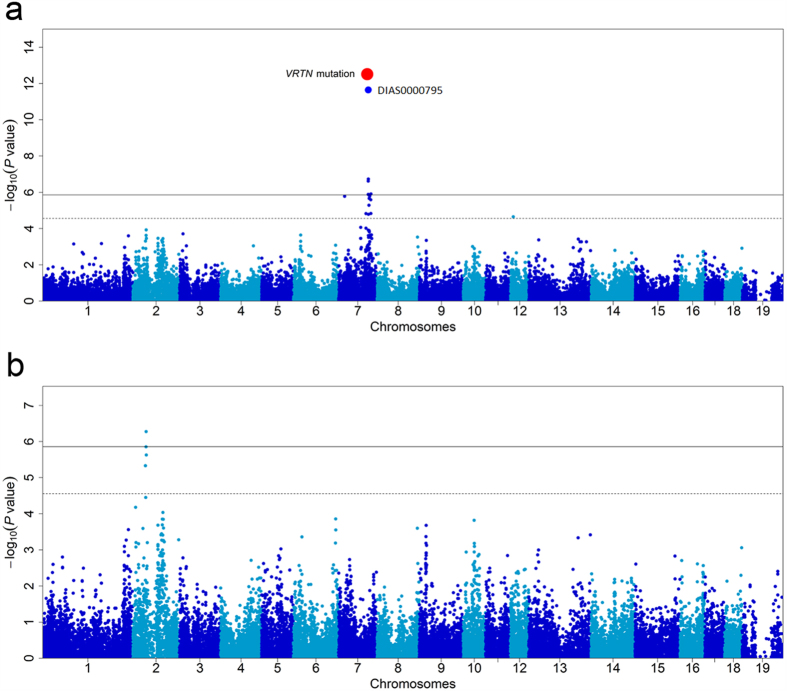
Figure 1. GWAS mapping for the number of thoracic vertebrae in the Erhualian population.
(a) Manhattan plots of the GWAS for the number of thoracic vertebrae in the Erhualian population. In the Manhattan plots, negative log10 P values of the quantified SNPs were plotted against their genomic positions. Different colors indicate different chromosomes. The red dot represents the VRTN mutation, and the top GWAS SNP (DLAS0000795) on the Illumina Porcine 60K Beadchips is indicated. The solid and dashed lines indicate the 5% genome-wide and chromosome-wide Bonferroni-corrected thresholds, respectively. (b) When the VRTN mutation was include as a fixed effect in the GWAS model, no other SNP on SSC7 showed association signal.