Abstract
In this study, we constructed a protein-protein interaction (PPI) network of B. licheniformis strain WX-02 with interolog method and domain-based method, which contained 15,864 edges and 2,448 nodes. Although computationally predicted networks have relatively low coverage and high false-positive rate, our prediction was confirmed from three perspectives: local structural features, functional similarities and transcriptional correlations. Further analysis of the COG heat map showed that protein interactions in B. licheniformis WX-02 mainly occurred in the same functional categories. By incorporating the transcriptome data, we found that the topological properties of the PPI network were robust under normal and high salt conditions. In addition, 267 different protein complexes were identified and 117 poorly characterized proteins were annotated with certain functions based on the PPI network. Furthermore, the sub-network showed that a hub protein CcpA jointed directly or indirectly many proteins related to γ-PGA synthesis and regulation, such as PgsB, GltA, GltB, ProB, ProJ, YcgM and two signal transduction systems ComP-ComA and DegS-DegU. Thus, CcpA might play an important role in the regulation of γ-PGA synthesis. This study therefore will facilitate the understanding of the complex cellular behaviors and mechanisms of γ-PGA synthesis in B. licheniformis WX-02.
Bacillus licheniformis (B. licheniformis) is a gram-positive spore-forming bacterium widely used in industry and agriculture1. For example, it can be used to produce many commercial enzymes2, biofuels and chemicals by fermentation, including poly-gamma-glutamic acid (γ-PGA)3, acetoin4 and antibiotics5, and even can be directly used to convert plumage into nutritious food for livestock6. Currently, the studies of B. licheniformis are mainly focused on one specific protein or several proteins in a single pathway7,8,9,10, while no comprehensive protein-protein interaction (PPI) network has been reported.
Proteins seldom perform their biological functions independently, and most complex cellular processes must be understood via large-scale PPI networks11,12. The availability of B. licheniformis strain WX-02 genome makes it possible to perform genome-scale analysis based on PPI network13,14. Genome-wide PPI networks have become powerful tools to study the cellular behaviors with a global view, and they can reveal the relationships between different kinds of proteins with various functions. Proteins involved in important biological processes and controlling the entire network can also be detected with the organization of the interactome11,15,16. In addition, the constructed PPI network is conducive to elucidating some protein functions that are poorly characterized with genome annotation17,18.
Currently, a large number of PPI networks have been constructed with high-throughput experimental methods, such as yeast two-hybrid system and tandem affinity purification19. However, these methods are quite costly in time and money20,21. With the increasing number of experimentally-determined PPIs and 3D-structures of proteins, a series of computational methods have been developed and attracted researchers by economical, rapid and convenient characters. In this study, we predicted the PPI network of B. licheniformis WX-02 by using two independent computational methods (interolog method and domain-based method) and analyzed the network from different perspectives. Finally, a PPI network containing 15,864 edges and 2,448 nodes was obtained. Based on this network, we investigated some species-specific properties of the network to explore the features of B. licheniformis WX-02 and dissected the functional modules related to γ-PGA biosynthesis to provide insights into its regulatory mechanism. The predicted PPI network can be used as a valuable resource for studying the physiology and metabolisms of B. licheniformis WX-02.
Results and Discussion
Construction of the genome-scale PPI network
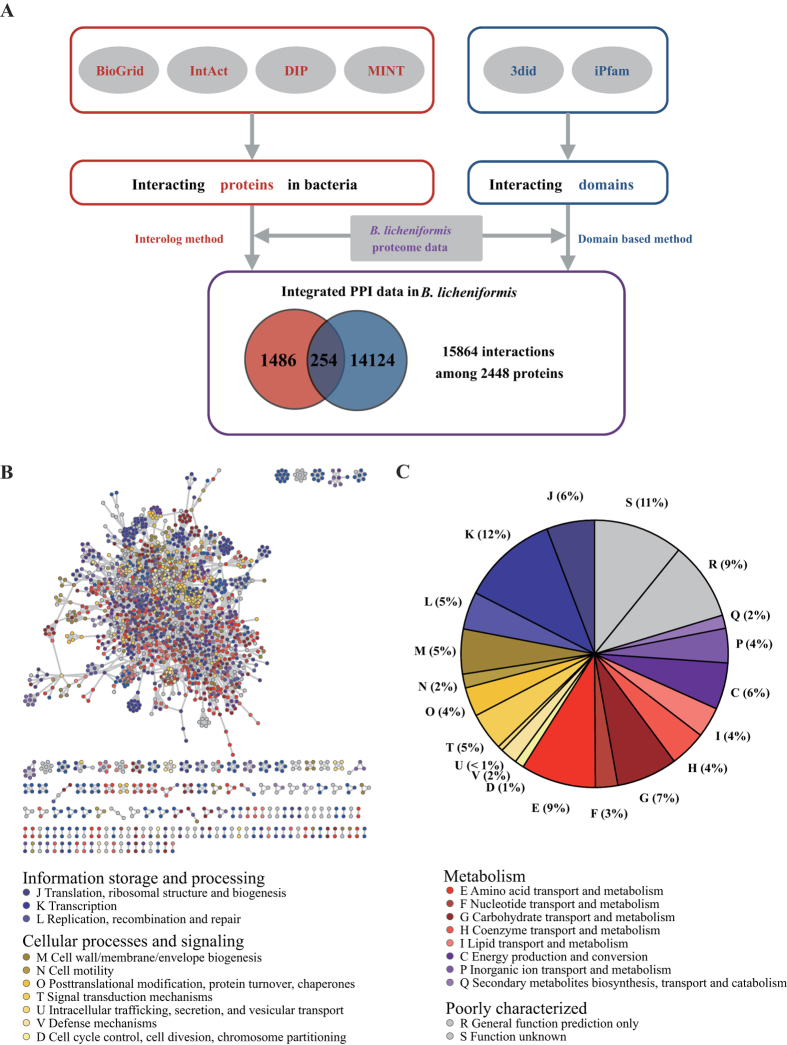
The PPI network was constructed by interolog method and domain-based method (Fig. 1A). These two methods predicted 1,740 and 14,378 PPIs respectively, and shared 254 PPIs. Finally, the merged non-redundant PPI network contains 15,864 edges and 2,448 nodes (see Supplementary Table S1 online). As homomeric interactions may cause bias in subsequent analysis, we excluded them from the network when investigating the relationships of interacting proteins22,23. As a result, the remained network comprised 13,664 interactions among 2,165 proteins.
Figure 1. Flowchart for constructing PPI network in B. licheniformis WX-02 and overview of the network.
(A) Flowchart for constructing PPI network. (B) Nodes of the network are colored according to their COG categories, and therefore nodes with the same color belong the same functional category. (C) The proportions of COG functional categories in the PPI network.
The network was visualized by Cytoscape24 and nodes were colored according to their cluster of orthologous groups (COG) functional categories (Fig. 1B). The distribution of COG in PPI network is shown in Fig. 1C. Proteins involved in ‘transcription (K)’ accounted for the largest proportion (12%), which are highlighted in deep blue; while the proteins related to ‘intracellular trafficking, secretion, and vesicular transport (U)’ accounted for the smallest proportion (less than 1%), which are marked with light yellow. The above results suggest that many transcriptional regulation processes in B. licheniformis can be performed through the PPI network, which is similar to some cases reported in Bacillus subtilis (B. subtilis)25,26.
Quality assessment of the PPI network
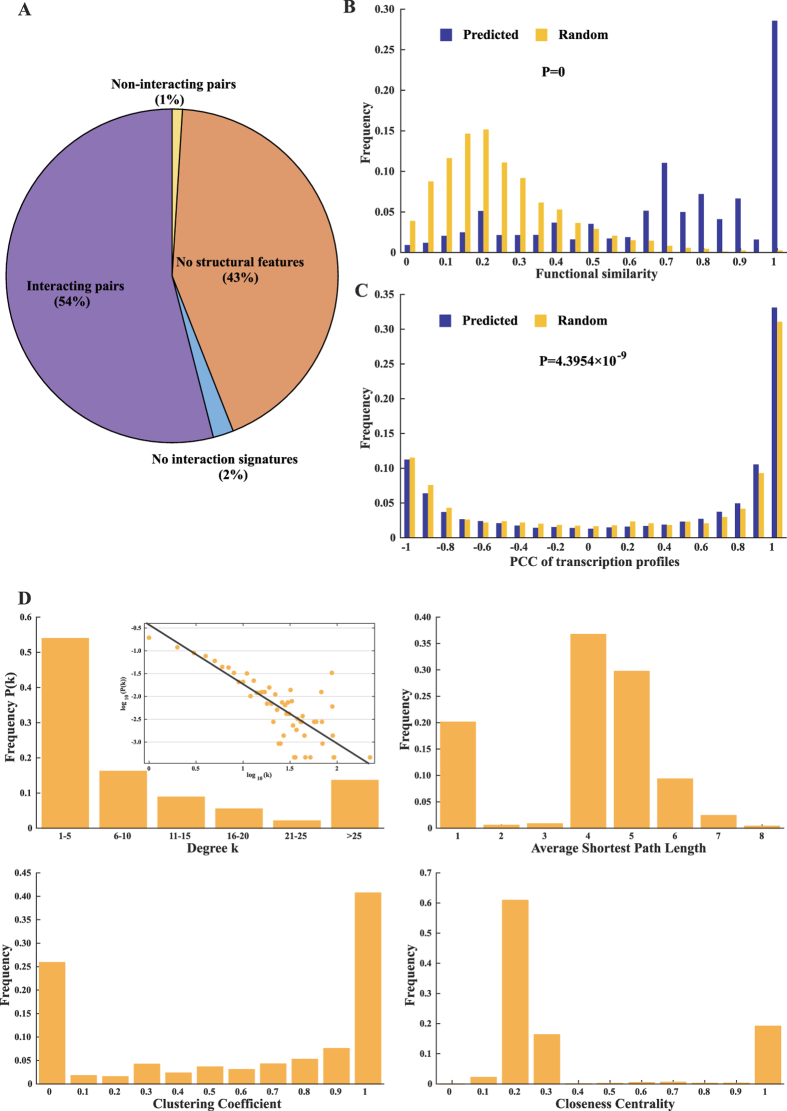
The accuracy of the predicted PPI network was evaluated from three perspectives: local structural features, functional similarities and gene transcription correlations. Firstly, we evaluated 1,000 randomly selected PPIs with a structural context method27,28. As well-characterized structural templates in available databases are limited, 43% of the selected PPIs contained at least one protein that had no structural features. Surprisingly, 54% of the PPIs could be confirmed and only 1% were classified as non-interacting pairs (Fig. 2A), indicating that more than half of our PPIs can be validated by local structural features and the PPI network is relatively reliable.
Figure 2. Validation and topological properties of the B. licheniformis WX-02 PPI network.
(A) 1,000 randomly selected PPIs validated by iLoop web server. (B) Comparison of the GO similarity between the predicted PPI network and random networks with same topology. (C) Comparison of the PCC of gene transcription profiles between protein pairs derived from the PPI network and random networks with same topology. (D) Topological properties.
Functional similarities of interacting proteins can also be used to evaluate the quality of PPIs, since interacting proteins are prone to have similar functions29,30. We calculated the functional similarities of protein pairs in the PPI network and in random networks with the same topology according to their semantic similarities of gene ontology (GO) annotations based on reference31. Figure 2B shows that the functional similarities of protein pairs in the PPI network (mainly falling within 0.65 ~ 1) are significantly higher than those in random networks (most of which are less than 0.4).
In addition, we compared the Pearson correlation coefficient (PCC) of normalized transcription profiles between interacting and random protein pairs. Previous studies have demonstrated that interacting proteins tend to have similar transcription patterns32. Hence, an accurate PPI network should contain significantly more interacting protein pairs with similar transcription patterns than random networks. Based on gene transcription, we calculated the PCC between protein pairs in the PPI network and those in random networks with the same topology, respectively. Figure 2C demonstrates that the PCC value of transcription profiles of protein pairs in the PPI network is significantly higher than that in random networks.
Despite the fact that the resolution of theoretical methods is lower than that of some structural modeling methods33,34, and the PPIs detected in our study do not cover all the actually existing PPIs, the above results indicate a high accuracy of the predicted B. licheniformis PPI network.
Properties of the PPI network
We calculated and analyzed the topological parameters of PPI network with Network Analysis plugins in Cytoscape24. As the case for many complex networks35, degree distribution of the PPI network in B. licheniformis WX-02 follows the power law, which characterizes the PPI network as a scale-free network (Fig. 2D). The average degree of this network is 12.6 and the degrees of 70% proteins are lower than 10. The average path length, cluster coefficient and the number of sub-networks are 4.7, 0.61 and 150, respectively. The largest sub-network contains 13,057 interactions and 1,718 proteins. Figure 2D shows that the distribution of average short path length, clustering coefficient and closeness centrality has two peaks, indicating the existence of many small sub-networks, whose topological parameters are quite different from those of the largest sub-network.
For the predicted PPI network, the degree exponent γ was calculated as 1.6 by the maximum likelihood estimate. It is well known that if the degree exponent is smaller than 2, relatively fewer nodes are needed to control the entire network36. These nodes were identified by minimum dominating set (MDS), since a previous study has reported that they play an important role in controlling the network16. In the present study, we determined a MDS in the B. licheniformis WX-02 PPI network by solving an integer-based linear programming problem. The resulting MDS contains 406 nodes, which account for less than 20% of the total nodes. To further analyze these important nodes, we performed COG enrichment analysis for them, finding that the proteins in MDS are significantly enriched in ‘carbohydrate transport and metabolism (G, fisher’s exact test, P < 0.05)’, ‘replication, recombination and repair (L, fisher’s exact test, P < 0.05)’ and ‘unknown function (S, fisher’s exact test, P < 0.01)’ (see Supplementary Table S2 online). Since the proteins in MDS are enriched in essential functional categories, such as cancer-related and virus-targeted genes in the PPI network of Homo sapiens and Saccharomyces cerevisiae16, the proteins with unknown function belonging to MDS in our PPI network might be involved in some important biological processes.
Heat map of COG functions in the PPI network
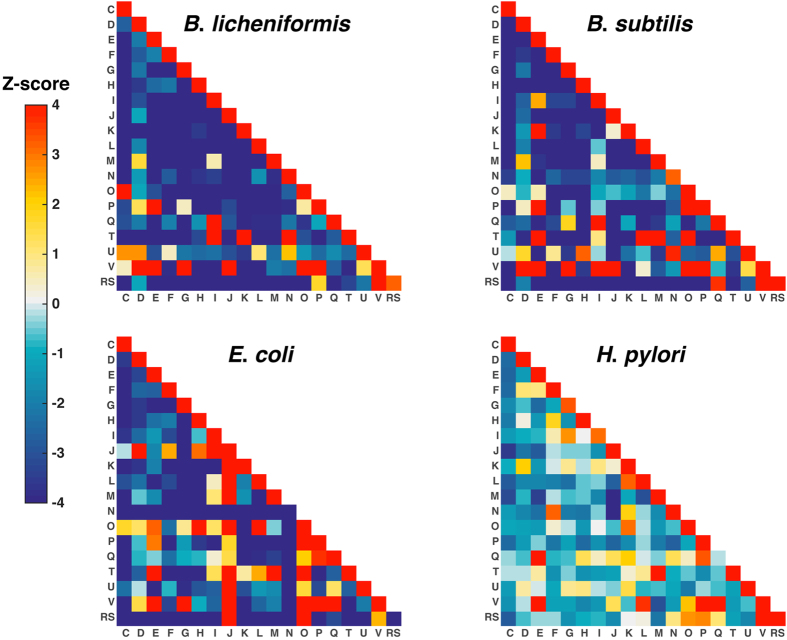
In this study, we performed PPI enrichment analysis by presenting the PPI network as a heat map based on different COG categories (Fig. 3)32,37,38,39. The PPI networks of other three model species (B. subtilis 168, E. coli K12 and H. pylori 26695) and their corresponding heat maps were constructed for comparison. To ensure the reliability of the comparative results, we used the same computational methods and reference PPI data to establish their PPI networks as B. licheniformis. Finally, the networks of B. subtilis, E. coli and H. pylori include 15,862, 23,900 and 2,965 PPIs, among which 15,304, 22,945 and 2,287 have COG annotations respectively. From Fig. 3, it can be observed that the PPI data of these four strains are mainly enriched in diagonal regions, suggesting that most of the interactions occur within the same functional categories.
Figure 3. Heat map of COG functional categories of PPIs for four organisms (B. licheniformis, B. subtilis, E. coli and H. pylori).
The numbers of PPIs among various COG functional categories were normalized by Z-score from a statistical model. The color indicates the enrichment degree of interactions between COG function categories.
Nevertheless, the differences among the four heat maps are obvious, indicating the species-specific functional features of these bacterial strains. In E. coli, the majority of PPIs are related to ‘translation, ribosomal structure and biogenesis (J)’ or ‘posttranslational modification, protein turnover, chaperones (O)’, while in the other three strains, most PPIs are not dominated by one or two classes of proteins. In Bacillus species, the proteins related to ‘defense mechanisms (V)’ tend to interact with the proteins from ‘Intracellular trafficking, secretion, and vesicular transport (U)’, while this phenomenon was not observed in other two gram-negative bacteria. Therefore, it can be speculated that Bacillus species might have specific defense mechanism to protect themselves. Moreover, several specific functional features were discovered in B. licheniformis WX-02. For instance, we found that the proteins in ‘signal transduction mechanisms (T)’ category are highly connected with those in ‘transcription (K)’ category and ‘cell motility (N)’ category. On the other hand, it is interesting that the interactions between ‘Cell wall/membrane/envelope biogenesis (M)’ and ‘Signal transduction mechanisms (T)’ proteins are all enriched in the networks of B. subtilis, E. coli and H. pylori, except for in that of B. licheniformis. These different features suggest that there might be unique complex regulatory mechanisms in B. licheniformis WX-02, which provide an effective way to explain its physiological characteristics and complex cellular behaviors.
Analysis and comparison of the PPI networks under normal and high salt conditions
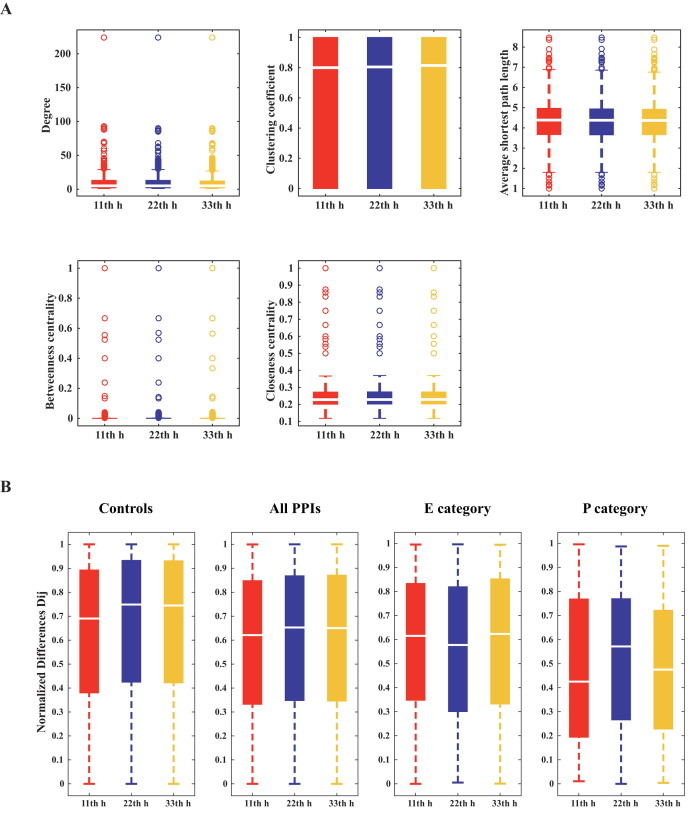
To investigate the dynamics of the PPI networks under normal and high salt conditions, we incorporated the strand-specific RNA-seq (ssRNA-seq) data into the PPI network and obtained three sub-networks with expressed genes at different time points (network1 for normal condition at 11th h, network2 for early long-term salt adaption at 22th h and network3 for late long-term salt adaption at 33th h)14. In order to explore the differences and similarities of these three networks, we performed analysis from two perspectives: topology and transcription differences between the interacting proteins. Firstly, we analyzed their topological properties by calculating 5 local topology metrics for each node in the corresponding networks, including degree, clustering coefficient, average shortest path length, betweenness centrality and closeness centrality. Interestingly, no significant differences were detected in the distributions of these local topology metrics for the three networks (Fig. 4A). These comparative results suggest that though the transcription levels and phenotypes are significantly different under normal and high salt conditions14, the topological properties of the PPI network are robust.
Figure 4. Comparison of local topological properties and normalized difference under normal and high salt conditions.
(A) Comparison of degree, clustering coefficient, average shortest path length, betweenness centrality and closeness centrality by Wilcoxon rank sum test. (B) Comparison of normalized differences for three time points and four groups of protein pairs. ‘Control’ represents all the pairwise proteins in the PPI network; ‘All PPI’ represents all the interacting protein pairs in the PPI network. ‘E category’ represents the interacting protein pairs belonging to ‘amino acid transport and metabolism (E)’; ‘P category’ represents the interacting protein pairs belonging to ‘inorganic ion transport and metabolism (P)’.
On the other hand, we investigated the absolute transcription levels between the interacting proteins, because their relative stoichiometrical amounts can affect productivity and efficiency. To this end, we defined the normalized transcription difference as the proportion of the difference value between the reads per kilobase of ORF per million mapped reads (RPKM) of two interacting proteins to the sum of their RPKM values40. Figure 4B shows the normalized difference distribution of the protein pairs at three time points for four groups i.e., ‘control’ group (all possible protein pairs in the network), ‘all PPI’ group (all interacting protein pairs in the network), sub-networks related to ‘amino acid transport and metabolism (E category)’ and ‘inorganic ion transport and metabolism (P category)’. From Fig. 4B, it is observed that the normalized difference distribution of ‘control’ group (all possible protein pairs in the network) is higher than that of ‘all PPI’ group (all interacting protein pairs in the network), revealing that the transcription levels of the interacting proteins are more approximate. By comparing the normalized difference distribution of PPI networks for three time points, we found that the median of the normalized difference distribution of network1 was smaller than that of network2 and network3 (Fig. 4B), demonstrating that the normalized difference distribution of PPIs is affected under high salt condition, which is consistent with the analysis of transcription profiles. Interestingly, sub-networks of ‘E category’ and ‘P category’ exhibit opposite trends. The normalized difference distribution of interactions between the proteins related to ‘E category’ is decreased at 22th h and then is restored to the normal level at 33th h. These changes might result in a more rational ratio of interacting proteins that are responsible for amino acid metabolism and acceleration of amino acid synthesis. However, the normalized difference distribution of interactions between the proteins related to ‘P category’ proteins is increased at 22th h relative to the normal condition. This change might contribute to the weakening of ion transport processes, the diminishing of ion-exchange amount and the maintaining of a stable osmotic pressure under long-term salt adaption. At 33th h, the transcription levels of many ‘P category’ proteins decrease to the levels under normal condition. The above results might explain the change of colony forming units (CFU), as the CFU decreased rapidly after the addition of 6% NaCl solution to the medium at 11th h, then the strain slowly resumed growth at about 22th h and the biomass reached almost the same level as in 11th h at 33th h14.
Identification of the protein complexes and prediction of the functions for uncharacterized proteins
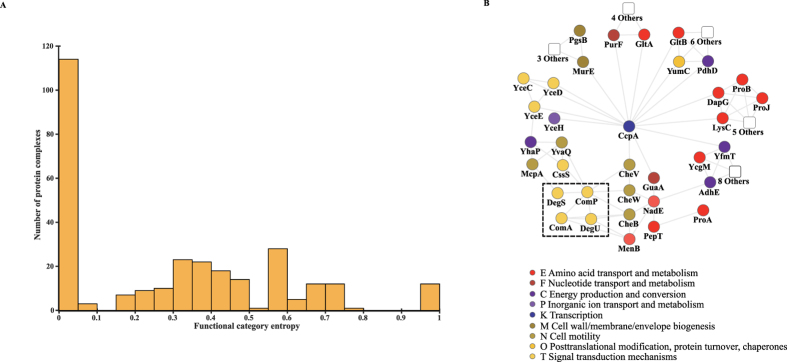
PPI network is a powerful tool to predict the functions of poorly characterized proteins. In this study, we proposed a two-step approach to determine the protein functions: identifying the protein complexes in the PPI network, and then predicting the protein functions based on these protein complexes. By using a clustering algorithm TSN-PCD41, we finally obtained 267 different protein complexes (see Supplementary Table S3 online). After obtaining the protein complexes, the functional category entropy for each protein complex was calculated according to COG functional categories. As expected, the functions of proteins belonging to the same protein complex are prone to be consistent (Fig. 5A), indicating that the protein function within a certain complex can be predicted through the enriched COG functional categories. With this module-assisted method42, we finally annotated 117 proteins with unknown functions (see Supplementary Table S4 online).
Figure 5. Functional category entropy distribution of protein complexes and sub-network related to γ-PGA synthesis and regulation.
(A) Functional category entropy distribution of protein complexes. (B) The proteins are colored based on their COG functional categories.
Analysis of the sub-network related to γ-PGA synthesis and regulation
Some studies have reported that B. licheniformis WX-02 can produce γ-PGA under normal condition and has a much higher yield under high salt environment14,43. Up to now, genes (pgsB, pgsC, pgsA and pgsE) related to γ-PGA synthesis have been reported in B. subtilis and B. licheniformis. Although a series of molecular and cellular studies have been performed on B. licheniformis, the regulation mechanism of γ-PGA is still not clear. Here, we analyzed the sub-network related to γ-PGA synthesis and regulation. Figure 5B shows that a hub protein CcpA directly or indirectly joints many proteins related to γ-PGA synthesis (PgsB) and regulation, such as GltA and GltB (which together encode glutamate-oxoglutarate amidotransferase), proteins related to proline metabolism (ProB, ProJ, YcgM) and two signal transduction systems ComP-ComA and DegS-DegU (Fig. 5B).
The CcpA transcriptional regulator is a central regulatory factor in the intersection between carbon and nitrogen metabolism44, and can regulate the metabolisms by interacting with other proteins45. According to Fig. 5B, it can be inferred that CcpA might also be related to γ-PGA synthesis through the PPI network. To illustrate this point, we further analyzed the sub-network. First of all, the γ-PGA synthesis protein PgsB can interact indirectly with CcpA through UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase (murE). Also, CcpA can interact directly with proteins GltA and GltB encoding glutamate-oxoglutarate amidotransferase (GOGAT), which play an important role in the upstream pathway of γ-PGA synthesis46. Thus, it can be speculated that CcpA might affect the γ-PGA synthesis by regulating the GOGAT through protein interactions. In addition, CcpA is connected with several chemotaxis proteins, and further interacts with two signal transduction systems ComP-ComA and DegS-DegU. It is well known that the synthesis of γ-PGA is under the control of these two signal transduction systems47,48. These results suggest that CcpA might first interact with chemotaxis proteins and regulate their expression, and then these chemotaxis proteins affect the regulation of ComP-ComA and DegS-DegU to regulate the γ-PGA synthesis of B. licheniformis WX-02. Based on the above analyses, CcpA can play an important central role in the regulation of γ-PGA synthesis through interacting with some related proteins.
Conclusions
In this work, we presented a genome-wide PPI network with 15,864 edges and 2,448 nodes of B. licheniformis WX-02 by combining interolog method and domain based method. The PPI network was subsequently verified from three perspectives: local structural features, functional similarities and transcription correlations. Although the predicted PPI network is far from perfect, it can provide new insights into the research of B. licheniformis WX-02. By analyzing and comparing the networks under normal and high salt conditions based on transcriptome data, we found that the topological properties of the PPI network are robust to tolerate fluctuations in transcription levels as well as changes in environmental conditions. In addition, we predicted 267 different protein complexes and annotated 117 poorly uncharacterized proteins based on the network. Further analyses of the sub-network show that the hub protein CcpA interacts directly or indirectly with many proteins involved in γ-PGA synthesis and regulation, indicating that CcpA might play an important role in regulating γ-PGA synthesis through the PPI network. The predicted PPI network will provide a significant foundation for exploring the molecular mechanisms of B. licheniformis WX-02 and developing optimized industry strains for producing chemicals.
Material and Methods
Data source
To construct the PPI network of B. licheniformis, we collected both the experimental interacting protein pairs and domain pairs from the databases. Totally, 44,648 experimental PPIs among 11,196 proteins for bacteria were downloaded from BioGRID49, IntAct50, DIP51 and MINT52 databases (Table 1). 9,590 domain-domain interactions (DDIs) among 5,619 domains were collected from iPfam53 and 3did54 databases. All the protein sequences were retrieved from NCBI RefSeq and UniProt. Domain alignment profiles were obtained from Pfam database55.
Table 1. High-quality PPIs and DDIs obtained from different public databases.
| Database | No. Proteins/Domains | No. PPIs/DDIs |
|---|---|---|
| BioGRID | 70 | 66 |
| IntAct | 10,232 | 38,758 |
| DIP | 4,259 | 14,595 |
| MINT | 1,384 | 3,782 |
| 3did | 5,466 | 8,651 |
| iPfam | 4,775 | 9,516 |
Interolog method
This prediction method is based on the conserved proteins in different species56. We detected the potential orthologs between B. licheniformis and reference organisms using BLASTP (E-value ≤ 10−5, sequence identity ≥30%, and alignment coverage ≥60%). To ensure the accuracy of the predicted results, protein pairs with the highest alignment score were kept if a protein corresponded to multiple homologs in one organism. This process might reduce the number of predicted interactions, but it could minimize the false positive rate. For any two proteins in B. licheniformis, if their orthologs in the reference genomes had at least one experimentally determined interaction, the two proteins were considered to have interaction.
Domain-domain interaction based method
The method attempts to predict protein interactions based on the experimentally and structurally determined DDIs. For a protein pair (X and Y) in B. licheniformis, we assumed that m and n were one domain in protein X and Y respectively. If m and n were proved to be an experimental interacting domain pair, X and Y were considered to have interaction. Domains of proteins in B. licheniformis were predicted based on the Pfam domain database and HMMER program (E-value ≤ 10−5, bias ≤ 1)57. The interacting domain pairs were checked based on the data from iPfam and 3did databases.
Network validation
To confirm the predicted PPIs, we randomly selected 1,000 PPIs and submitted them to the PPI prediction web server (http://sbi.imim.es/iLoops.php)28. This web server, which defines protein structural features based on the loops from ArchDB58 and domains from SCOP59, was used to validate the PPIs by evaluating whether loop or domain patterns from two input proteins had interaction signatures with random forest classifier.
In addition, we used a method based on GO functional similarities to confirm the PPIs. It is well known that two interacting proteins tend to have similar or related functions. Based on this assumption, we compared the GO functional similarities between the predicted PPI network and 100 random networks (with the same topology as the PPI network). The GO annotations of B. licheniformis genome were downloaded from GO database60. Totally, 1,682 of 2,165 proteins in the predicted PPI network had GO annotation. Then, the semantic similarities of GO terms and functional similarities of proteins in the PPI and random networks were calculated with the algorithms proposed by reference31. The comparison of functional similarity distributions between PPI network and random networks was performed with Wilcoxon rank-sum test.
Moreover, gene transcription correlations of interacting proteins were also used to access the reliability of the PPI network. The ssRNA-seq data of B. licheniformis WX-02 for three time points (11th h, 22th h and 33th h) were obtained from the previous study14. The PCC of gene transcription profiles of the protein pairs in the PPI and 100 random networks was compared. The statistical difference between the predicted PPI network and random networks was also measured by P-value from Wilcoxon rank-sum test.
Analysis of COG functional heat map
Based on COG functional categories, the PPI data were presented as heat map. Colors in the heat map indicate Z-scores calculated by a statistical model. Considering that a randomized network contained same nodes as the predicted PPI network, the probability for a protein in functional class i to interact with a protein in functional class j in the randomized network was calculated as:
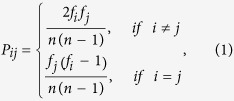 |
where n is the total number of proteins in the predicted PPI network and fi is the number of proteins belonging to functional class i. In the randomized network, the number of interactions between proteins from functional class i and j was assumed to follow a binomial distribution. Finally, the Z-scores were calculated as:
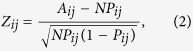 |
where Aij, NPij and NPij(1 − Pij) represent the actual value, expected value and variance of the number of interactions between proteins from functional class i and j, respectively.
Dynamic changes of the PPI network under normal and high salt conditions
The transcription profiles were obtained from three sample points: 11th h (0 h after the onset of 6% NaCl), 22th h (11 h after the onset of exposure to 6% NaCl) and 33th h (22 h after the onset of exposure to 6% NaCl), which have been reported in the previous study14. Firstly, we used RPKM to represent the normalized transcription levels of genes. Then, we assigned the RPKM values of each time point to the corresponding nodes in PPI network to obtain three networks. Here we defined a rule: if the RPKM value of a gene was lower than 1, this gene was considered to have no effect on the PPI network and would be removed from the network. Based on this process, we obtained three new PPI networks (network1 for 11th h, network2 for 22th h and network3 for 33th h) for different experimental conditions.
We calculated the normalized transcription difference Dij between a pair of proteins i and j as defined in the previous study40:
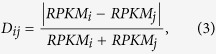 |
where RPKMi represents the RPKM value of gene i, and this value ranges from 0 to 1.
Prediction of the protein complexes and annotation of the protein functions
Protein complexes in the PPI network were identified using a clustering algorithm named TSN-PCD41. The inputs of TSN-PCD were PPIs and gene transcription data, which were used to generate time-series sub-networks. Then the clustering was performed based on these subnetworks. In this algorithm, the threshold of gene transcription level (RPKM value), λ (a parameter affecting the clustering results) and size value were set as 1, 1 and 3, respectively. In information theory, entropy is used to measure uncertainty or variability of complex systems. In this study, we defined the functional category entropy of a protein complex to indicate the function homogeneity of the protein complex. The functional category entropy was calculated as follows:
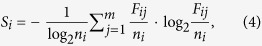 |
where ni is the number of proteins in the complex i and Fij is the number of proteins annotated with the function j in the complex i. The lower entropy means greater homogeneity. The homogeneity is ascribed to a specific function enriched in a protein complex. Therefore, we could assign functions to the uncharacterized proteins with the functions enriched in a protein complex. The function enrichment analysis was performed based on fisher’s exact test.
Additional Information
How to cite this article: Han, Y.-C. et al. Prediction and characterization of protein-protein interaction network in Bacillus licheniformis WX-02. Sci. Rep. 6, 19486; doi: 10.1038/srep19486 (2016).
Supplementary Material
Acknowledgments
This research was supported by National Natural Science Foundation of China (31271406 and 31071659) and the program for New Century Excellent Talents in University (NCET-13-0807).
Footnotes
Author Contributions Conceived and designed the experiments: L.L.C., J.G. and Y.C.H. Performed the experiments: J.G., Y.C.H., J.M.S., L.W. and C.C.S. Analyzed the data: J.G., Y.C.H., J.M.S., L.W. and C.C.S. Contributed reagents/materials/analysis tools: J.G., Y.C.H., J.M.S., L.W. and C.C.S. Wrote the paper: L.L.C., J.G. and Y.C.H. Wrote the script used for data analysis: J.G., Y.C.H., J.M.S., L.W. and C.C.S.
References
- Pötter M., Oppermann-Sanio F. B. & Steinbüchel A. Cultivation of bacteria producing polyamino acids with liquid manure as carbon and nitrogen source. Appl. Environ. Microbiol. 67, 617–622 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veith B. et al. The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J. Mol. Microbiol. Biotechnol. 7, 204–211 (2004). [DOI] [PubMed] [Google Scholar]
- Konglom N., Chuensangjun C., Pechyen C. & Sirisansaneeyakul S. Production of poly-γ-glutamic acid by Bacillus licheniformis, synthesis and characterization. Journal of Metals, Materials and Mineral 22, 7–11 (2012). [Google Scholar]
- Liu Y. F. et al. Efficient production of acetoin by the newly isolated Bacillus licheniformis strain MEL09. Process Biochem. 46, 390–394 (2011). [Google Scholar]
- McInerney M. J., Javaheri M. & Nagle D. P. Jr. Properties of the biosurfactant produced by Bacillus licheniformis strain JF-2. J. Ind. Microbiol. 5, 95–101 (1990). [DOI] [PubMed] [Google Scholar]
- Burtt E. H. & Ichida J. M. Occurrence of feather-degrading bacilli in the plumage of birds. Auk 116, 364–372 (1999). [Google Scholar]
- Liang C. et al. Enhancement of L-valine production in Bacillus licheniformis by blocking three branched pathways. Biotechnol. Lett. 37, 1243–1248 (2015). [DOI] [PubMed] [Google Scholar]
- Tian G. et al. Enhanced expression of pgdS gene for high production of poly-γ-glutamic aicd with lower molecular weight in Bacillus licheniformis WX-02. J. Chem. Technol. Biot. 89, 1825–1832 (2014). [Google Scholar]
- Qiu Y., Xiao F., Wei X., Wen Z. & Chen S. Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Appl. Microbiol. Biotechnol. 98, 8895–8903 (2014). [DOI] [PubMed] [Google Scholar]
- Qi G. et al. Deletion of meso-2, 3-butanediol dehydrogenase gene budC for enhanced D-2, 3-butanediol production in Bacillus licheniformis. Biotechnol. Biofuels 7, 16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor I. W. et al. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat. Biotechnol. 27, 199–204 (2009). [DOI] [PubMed] [Google Scholar]
- Rual J. F. et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 (2005). [DOI] [PubMed] [Google Scholar]
- Yangtse W. et al. Genome sequence of Bacillus licheniformis WX-02. J. Bacteriol. 194, 3561–3562 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. et al. Comprehensive transcriptome and improved genome annotation of Bacillus licheniformis WX-02. FEBS Lett. 589, 2372–2381 (2015). [DOI] [PubMed] [Google Scholar]
- Han J. D. et al. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature 430, 88–93 (2004). [DOI] [PubMed] [Google Scholar]
- Wuchty S. Controllability in protein interaction networks. Proc. Natl. Acad. Sci. USA 111, 7156–7160 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwikowski B., Uetz P. & Fields S. A network of protein-protein interactions in yeast. Nat. Biotechnol. 18, 1257–1261 (2000). [DOI] [PubMed] [Google Scholar]
- Mostafavi S., Ray D., Warde-Farley D., Grouios C. & Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 9, S4 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman K. Construction and analysis of protein-protein interaction networks. Automot. Exp. 2, 2 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E. M. & Fields S. Protein-protein interactions methods for detection and analysis. Microbiol. Rev. 59, 94–123 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C. et al. Comparative assessment of large-scale data sets of protein protein interactions. Nature 417, 399–403 (2002). [DOI] [PubMed] [Google Scholar]
- Mrowka R., Patzak A. & Herzel H. Is there a bias in proteome research? Genome Res. 11, 1971–1973 (2001). [DOI] [PubMed] [Google Scholar]
- Mrowka R., Liebermeister W. & Holste D. Does mapping reveal correlation between gene expression and protein–protein interaction? Nat. Genet. 33, 16–17 (2003). [DOI] [PubMed] [Google Scholar]
- Shannon P. et al. Cytoscape a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray L. V., Zalieckas J. M. & Fisher S. H. Bacillus subtilis glutamine synthetase controls gene expression through a protein-protein interaction with transcription factor TnrA. Cell 107, 427–435 (2001). [DOI] [PubMed] [Google Scholar]
- Commichau F. M., Herzberg C., Tripal P., Valerius O. & Stülke J. A regulatory protein–protein interaction governs glutamate biosynthesis in Bacillus subtilis: the glutamate dehydrogenase RocG moonlights in controlling the transcription factor GltC. Mol. Microbiol. 65, 642–654 (2007). [DOI] [PubMed] [Google Scholar]
- Planas-Iglesias J. et al. Understanding protein–protein interactions using local structural features. J. Mol. Biol. 425, 1210–1224 (2013). [DOI] [PubMed] [Google Scholar]
- Planas-Iglesias J., Marin-Lopez M. A., Bonet J., Garcia-Garcia J. & Oliva B. iLoops: a protein–protein interaction prediction server based on structural features. Bioinformatics 29, 2360–2362 (2013). [DOI] [PubMed] [Google Scholar]
- Lehner B. & Fraser A. G. A first-draft human protein-interaction map. Genome Biol. 5, R63 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häuser R. et al. A second-generation protein-protein interaction network of Helicobacter pylori. Mol. Cell Proteomics 13, 1318–1329 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. Z., Du Z., Payattakool R., Yu P. S. & Chen C. F. A new method to measure the semantic similarity of GO terms. Bioinformatics 23, 1274–1281 (2007). [DOI] [PubMed] [Google Scholar]
- Ge H., Liu Z., Church G. M. & Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat. Genet. 29, 482–486 (2001). [DOI] [PubMed] [Google Scholar]
- Mosca R., Pons T., Céol A., Valencia A. & Aloy P. Towards a detailed atlas of protein–protein interactions. Curr. Opin. Struc. Biol. 23, 929–940 (2013). [DOI] [PubMed] [Google Scholar]
- Szilagyi A. & Zhang Y. Template-based structure modeling of protein–protein interactions. Curr. Opin. Struc. Biol. 24, 10–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabási A. L. Scale-free networks: a decade and beyond. Science 325, 412–413 (2009). [DOI] [PubMed] [Google Scholar]
- Nacher J. C. & Akutsu T. Dominating scale-free networks with variable scaling exponent: heterogeneous networks are not difficult to control. New J. Phys. 14 (2012). [Google Scholar]
- Titz B. et al. The binary protein interactome of Treponema pallidum-the syphilis spirochete. PLoS One 3, e2292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peregrín-Alvarez J. M., Xiong X., Su C. & Parkinson J. The modular organization of protein interactions in Escherichia coli. PLoS Comput. Biol. 5, e1000523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Global protein-protein interaction network in the human pathogen Mycobacterium tuberculosis H37Rv. J. Proteome Res. 9, 6665–6677 (2010). [DOI] [PubMed] [Google Scholar]
- Jansen R., Greenbaum D. & Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 12, 37–46 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Wu X., Wang J. & Pan Y. Towards the identification of protein complexes and functional modules by integrating PPI network and gene expression data. BMC Bioinformatics 13, 109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharan R., Ulitsky I. & Shamir R. Network-based prediction of protein function. Mol. Syst. Biol. 3, 88 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Ji Z. & Chen S. Isolation of halotolerant Bacillus licheniformis WX-02 and regulatory effects of sodium chloride on yield and molecular sizes of poly-γ-glutamic acid. Appl. Biochem. Biotechnol. 160, 1332–1340 (2010). [DOI] [PubMed] [Google Scholar]
- Sonenshein A. L. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927 (2007). [DOI] [PubMed] [Google Scholar]
- Wünsche A. et al. CcpA forms complexes with CodY and RpoA in Bacillus subtilis. FEBS J. 279, 2201–2214 (2012). [DOI] [PubMed] [Google Scholar]
- Krog A., Heggeset T. M., Ellingsen T. E. & Brautaset T. Functional characterization of key enzymes involved in L-glutamate synthesis and degradation in the thermotolerant and methylotrophic bacterium Bacillus methanolicus. Appl. Environ. Microbiol. 79, 5321–5328 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S. P., Nagai T. & Itoh Y. Divergent structure of the ComQXPA quorum-sensing components: molecular basis of strain-specific communication mechanism in Bacillus subtilis. Mol. Microbiol. 37, 1159–1171 (2000). [DOI] [PubMed] [Google Scholar]
- Ohsawa T., Tsukahara K. & Ogura M. Bacillus subtilis response regulator DegU is a direct activator of pgsB transcription involved in γ-poly-glutamic acid synthesis. Biosci. Biotechnol. Biochem. 73, 2096–2102 (2009). [DOI] [PubMed] [Google Scholar]
- Stark C. et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 34, D535–D539 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H. et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 32, D452–D455 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I. et al. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 30, 303–305 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A. et al. MINT: the Molecular INTeraction database. Nucleic Acids Res. 35, D572–D574 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Miller B. L., Clements J. & Bateman A. iPfam: a database of protein family and domain interactions found in the Protein Data Bank. Nucleic Acids Res. 42, D364–D373 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosca R., Céol A., Stein A., Olivella R. & Aloy P. 3did: a catalog of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 42, D374–D379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. et al. The Pfam protein families database. Nucleic Acids Res. 32, D138–D141 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews L. R. et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 11, 2120–2126 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Clements J. & Eddy S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espadaler J. et al. ArchDB: automated protein loop classification as a tool for structural genomics. Nucleic Acids Res. 32, D185–D188 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva A. et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 36, 419–425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M. A. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–D261 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.