Figure 2. Protein structure context of natural helicase variants.
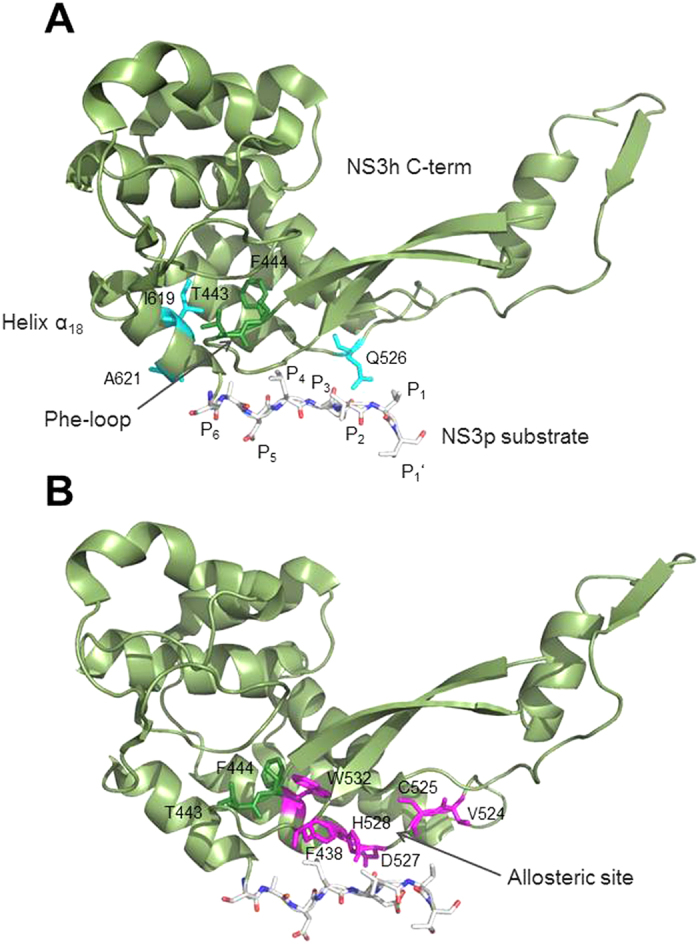
Details of PDB structure 1CU1 showing the C-terminal region of the NS3 helicase (amino acid residues 241 – 631). The NS3 protease natural substrate of proteolysis is given as a stick model with residues denoted as P6-P5-P4-P3-P2-P1-P1’ according to the nomenclature of Schechter and Berger32, in which residues surrounding a cleavage site are designated from the N- to the C-terminus with the scissile bond located between P1 and P1’. (A) Residues highlighted in cyan where mutations showed increased replication capacity. (B) Residues highlighted in magenta where mutations showed decreased replication capacity. Phe-loop residues T443 and F444 are highlighted in green.
