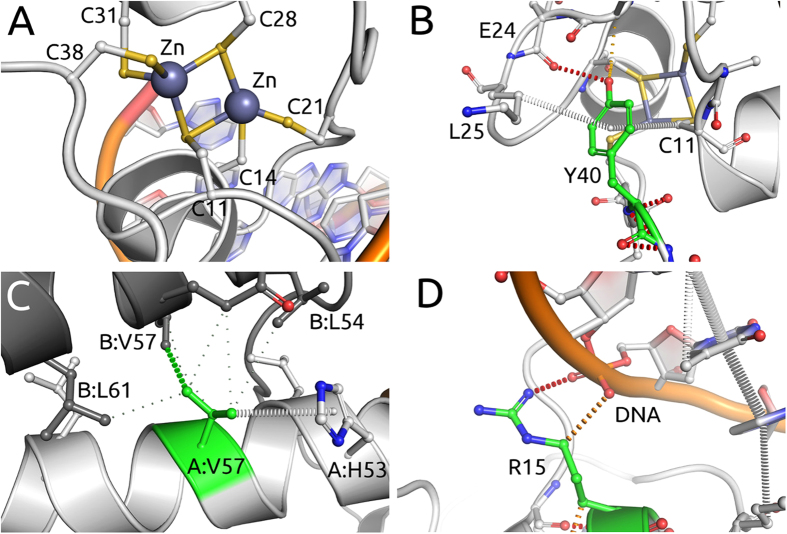
Figure 5. Noncovalent interaction networks in Gal4.
(A) depicts interactions between a pair of Zn2+ ions coordinated by six conserved cysteine residues in the Gal4-DNA complex. Mutations on these residues would, therefore, disrupt zinc binding, affecting Gal4 function. (B,C) depicts noncovalent interaction network of mutated residues in the Gal4-DNA complex. Mutated residues are depicted in green. Hydrogen bonds are depicted as red dotted lines, hydrophobic interactions in green and ring-ring interactions in grey. (Panel B) shows residue Tyr40 performing a side-chain to main-chain hydrogen bond and ring interactions. The mutation Y40A is predicted to be highly destabilizing, given the removal of a large portion of the side chain and consequent loss of interactions. (Panel C) shows residue Val57, whose mutations are predicted to also affect protein-protein affinity. Val57 establishes a network of hydrophobic and ring interactions that would be lost by the introduction of larger or hydrophilic residues, destabilizing the homodimer. (Panel D) shows residue R15, whose mutations are predicted to also affect RNA binding affinity. Arg15 directly interacts with the DNA molecule through a weak polar interaction and hydrogen bond. Mutations to aspartic and glutamic acids, through the introduction of an opposite charge, are predicted to destabilize the region and reducing DNA affinity.