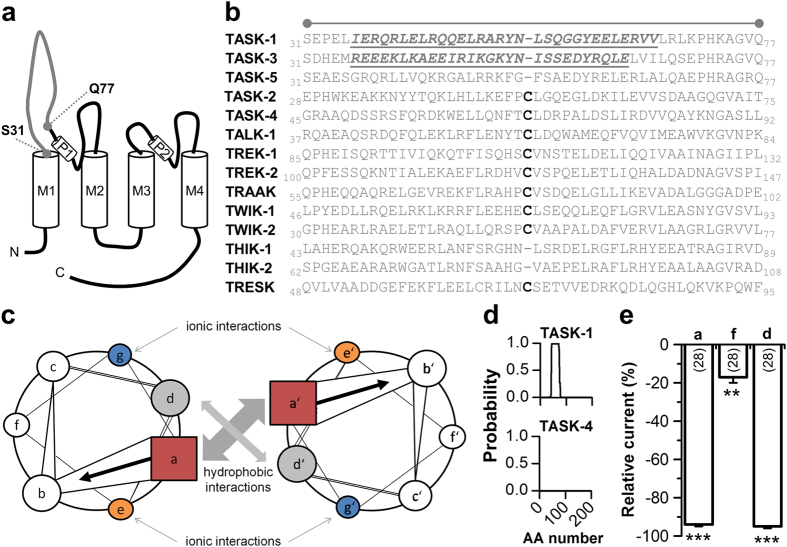
Figure 1. A coiled-coil prediction in the M1-P1 linker of TASK-1 channels.
(a) Transmembrane topology of TASK-1. The extracellular region corresponding to the cap structure of other K2P channels is indicated. (b) Amino acid sequence alignment of different K2P channels for the extracellular region indicated in (a). The cysteine residues, conserved in most K2P channels, are highlighted in bold. Residues of the four heptad repeat coiled-coil predictions in TASK-1 and TASK-3 are indicated in italics and by underlining. (c) Coiled-coil structures usually contain a repeated pattern of hydrophobic (h) and charged (c) amino-acid residues. ‘hxxhcxc’ refers to a heptad repeat re-occuring after every two turns of the helix. The positions within the heptad repeat are commonly labeled as ‘a–g’, where ‘a’ (red) and ‘d’ (gray) are the hydrophobic core positions, often occupied by the amino acids isoleucine, leucine, or valine, thus stabilizing helix dimerization through hydrophobic and van-der-Waals interactions, whereas ‘e’ (orange) and ‘g’ (blue) are typically solvent-exposed, polar residues (e.g. glutamate or lysine) that give specificity between the two helices through interhelical electrostatic interactions. The remaining three positions (‘b’, ‘c’, and ‘f’, white) must be all hydrophilic, as these will form helical solvent exposed surfaces. (d) Coiled-coil prediction tools revealed with a high probability a four-fold coiled-coil repeat in TASK-1 (top), whereas for instance in TASK-4 no coiled-coil domain was predicted (bottom). AA, amino acid. (e) Typical interaction sites of coiled-coil domains were mutated and current amplitudes, analyzed at +40 mV, were compared to wild-type TASK-1. ‘a’, indicates the four-fold mutation of all a-sites (R38A/Q45A/Y52A/Y59A); ‘f’, indicates the four-fold mutation of all f-sites (L43A/A50V/G57A/R64A); ‘d’, indicates the four-fold mutation of all d-sites (L41A/L48A/S55A/L62A). **p < 0.01; ***p < 0.001. Unpaired Student’s T-Test. Data are presented as Mean ± SEM. The number of experiments are included in the bar graph.