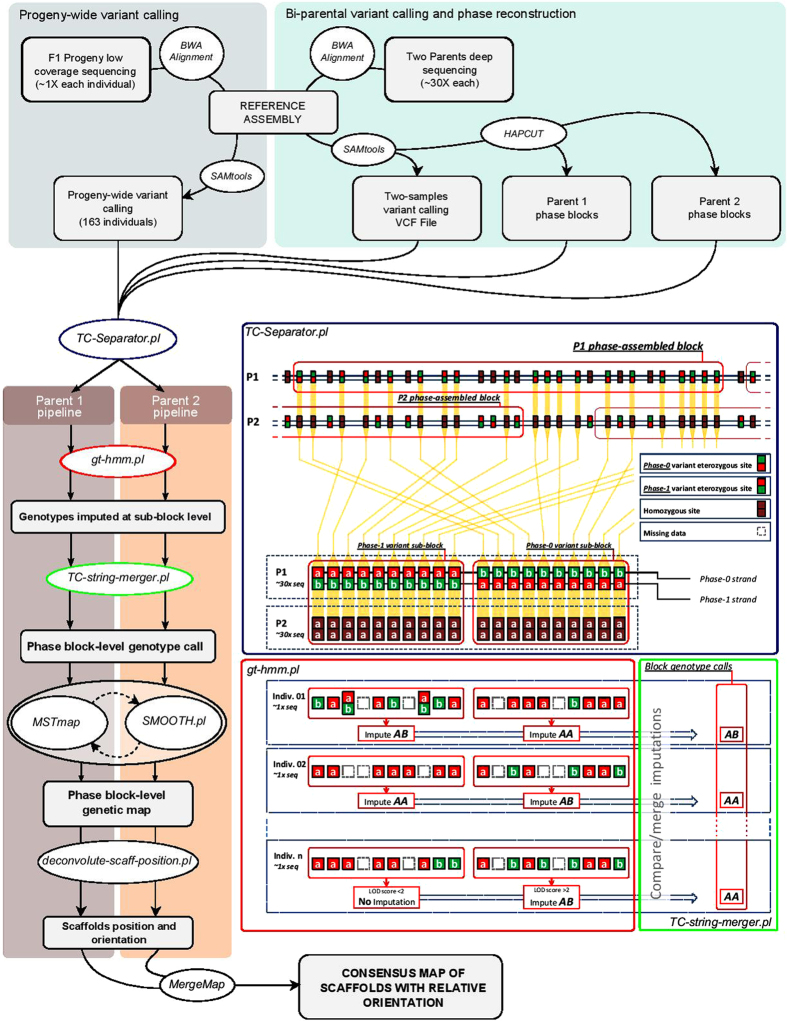
Figure 1. Workflow schema for the SoiLoCo pipeline used to anchor the C. cardunculus scaffolds in chromosomal pseudomolecules.
Alignments of parental reads to the draft scaffolds were used to (i) identify potential heterozygous test-cross sites and (ii) to compute haplotype phases in both parents (P1 and P2). A multi-sample VCF file of all the progeny was then processed to identify informative heterozygous sites based on parental SNPs and the phase of haplotype blocks (TC-separator.pl, blue box). This assigned the sites according to which phase (i.e. homologous chromosomes) they are expected to segregate in. Subsequently, an HMM-based algorithm was used to impute the most likely genotypes of each haplotype block segregating in the progeny (gt-hmm.pl, red box). A LOD score was also calculated to permit filtering of ambiguous imputations. Genotype imputation from the two alternative segregating phases were then summarized; when there was a discordant call between phases, a majority rule was applied and the highest LOD score for each segregating haplotype used to impute the most likely genotype (TC-string-merger.pl, green box). After grouping markers, linkage maps were generated for each parent using reiterative ordering with the MSTmap software (http://alumni.cs.ucr.edu/~yonghui/mstmap.html) and error correction using a Perl implementation of SMOOTH algorithm37. Maps were finally merged to generate a consensus map and to maximize the resolution of the order and orientation of scaffolds in chromosomal pseudomolecules.