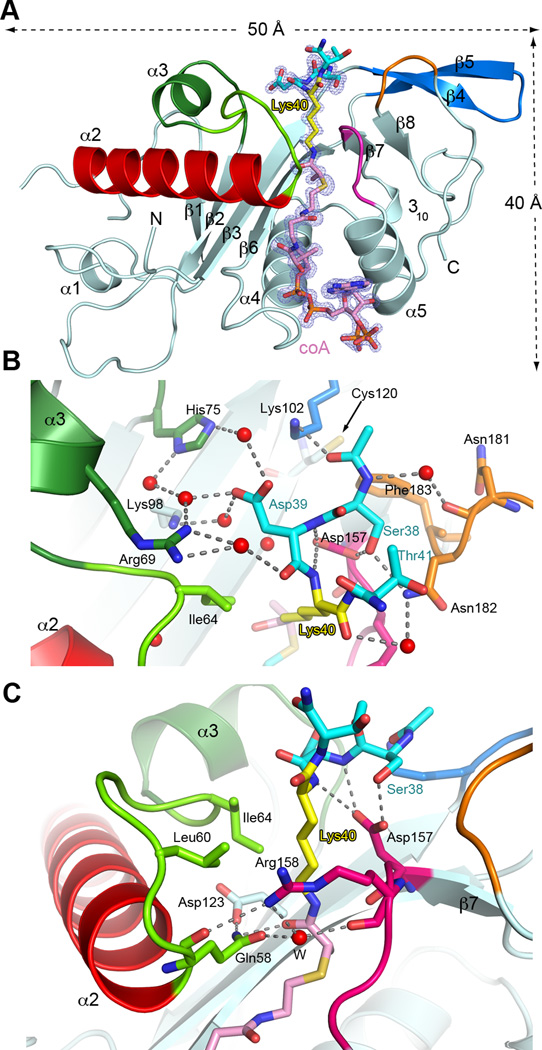
Figure 1.
Active site architecture and α−tubulin Lys40 loop recognition (A) Ribbon representation of TAT bound to peptide-coA bisubstrate analog 1 (IC50 ∼ 100 µM; Figures S1C and S1D); analog, shown as a stick model with Lys40 yellow, the rest of the peptide moiety, cyan, coA and linker, pink in the same scheme as Figure S1C; the |Fo|-|Fc| density (prior to modeling the ligand) is contoured at 3.5σ (blue); Regions engaged in substrate binding and catalysis colored green, red, magenta, orange and blue (B) Close-up of the active site showing residues engaged in tubulin peptide recognition; red spheres and dashed lines denote water molecules and hydrogen bonds, respectively; color scheme as in A; TAT residues labeled in black, peptide residues in color (C) Close-up of the active site showing residues important for Lys40 recognition and catalysis; colored as in A. Water molecule depicted as a red sphere labeled W. See also Figure S1 and Table S1.