Abstract
Background
In this study we report the genetic characterization, including expression analysis, of the genes involved in the uptake (NGT1) and catabolism (HXK1/NAG5, DAC1/NAG2, NAG1) of the aminosugar N-acetylglucosamine (GlcNAc) in Candida africana, a pathogenic biovariant of Candida albicans that is naturally unable to assimilate the GlcNAc.
Results
DNA sequence analysis of these genes revealed a number of characteristic nucleotide substitutions including a unique and distinctive guanine insertion that shifts the reading frame and generates a premature stop codon (TGA) 154 bp downstream of the ATG start codon of the HXK1 gene encoding the GlcNAc-kinase, a key enzyme of the GlcNAc catabolic pathway. However, all examined genes produced transcripts even though different levels of expression were observed among the Candida isolates examined. In particular, we found an HXK1-idependent relationship of the NGT1 gene and a considerable influence of the GlcNAc-kinase functionality on the transcription of the DAC1 and NAG1 genes. Additional phenotypic analysis revealed that C. africana isolates are hyperfilamentous in the first 24-48h of growth on filament-inducing media and revert to the yeast morphological form after 72h of incubation on these media.
Conclusions
Our results show that C. africana is a natural HXK1 mutant, displaying a number of phenotypic characteristics distinct from typical C. albicans isolates.
Introduction
Candida africana is a pathogenic fungus reported to cause genital infections in humans [1, 2]. It was first isolated in 1995 in Africa and subsequently proposed as a new Candida species closely related to the well-known fungal pathogen Candida albicans [3]. However, although C. africana exhibits a number of phenotypic characteristics clearly distinct from those observed in typical C. albicans isolates [3, 4], several molecular studies have demonstrated that these two fungi are, genetically, too closely related to be considered two distinct species [5, 6, 7, 8, 9]. Therefore, C. africana is currently considered to be a biovariant of C. albicans rather than a new species [1, 10].
Phenotypically, like C. albicans, C. africana produces characteristic germ-tubes in the presence of serum, however, unlike C. albicans, C. africana is unable to form chlamydospores and is unable to assimilate certain organic compounds including the two aminosugars N-acetylglucosamine and glucosamine [3].
N-acetylglucosamine (GlcNAc) is a monosaccharide essential for various cellular processes in both eukaryotic and prokaryotic organisms [11] and it is metabolized by 100% of typical C. albicans isolates [12]. In addition to utilizing GlcNAc as a carbon and energy source, in C. albicans GlcNAc also plays an important role in cell signaling by inducing white-opaque switching, the conversion between two heritable phenotypic states [13] and by modulating the yeast-to-hyphal transition [14] both of which are associated with virulence and pathogenicity in this fungus [15].
A previous proteomics study of plasma membrane proteins in C. albicans identified a unique and specific transporter (called NGT1) that allows for the uptake of GlcNAc in this species [16]. Once inside the cell, GlcNAc is phosphorylated, deacetylated and deaminated by three enzymes, encoded by the genes HXK1, DAC1 and NAG1, respectively. These enzymes ultimately convert GlcNAc into fructose-6-phosphate, which is used as an energy source by the cell [11].
C. africana is a natural GlcNAc-mutant, unable to use this monosaccharide for growth [3]. Furthermore, this fungus also shows a remarkably low level of filamentation [17], poor adhesion to human cells [18] and decreased virulence using Galleria mellonella as an infection model [17]. Since these properties are all linked to the ability to undergo morphological transitions, which in certain conditions is, in part, dependent on extracellular GlcNAc, we decided to study the sequence and expression levels of the genes involved in the GlcNAc catabolic pathway (also called NAG regulon) [14], including the NGT1 gene [16], in order to understand GlcNAc utilization in C. africana.
Materials and Methods
Fungal strains
Two C. africana strains: UPV/EHU 97135 from Bilbao, Spain [19] and CBS11016 from Centraalbureau voor Schimmelcultures, isolated in Vibo Valentia, Italy [8] were examined in this study. The identity of the strains was confirmed by partial amplification and detection of the HWP1 gene according to previous studies [20]. The C. albicans ATCC10231 strain was used as a reference strain in all experiments.
Ten Chinese C. africana strains (2313, 2286, 8513, 8840, 8866, 8350, 8367, E373, 5006, 5344 [21]) and one from Germany (A1622 [3]) were also included and used to confirm the observed genetic polymorphisms.
Phenotypic tests
To determine the extent to which C. africana is unable to grow in the presence of GlcNAc, all isolates were initially grown in liquid yeast nitrogen base (YNB) medium (Difco, Milan, Italy) containing 2% of galactose and then incubated at 30°C in the presence of different concentrations of GlcNAc (ranging from 0.195–100 mM). Assimilation tests were carried out as outlined in Kurtzman and Fell [22] using 5 ml of liquid YNB containing GlcNAc (first tube 100 mM) serially diluted to 0.195 mM. The test tubes were inoculated with 0.1 ml of a standardized fungal suspension (~107cells/ml) prepared spectrophotometrically, and incubated at 30°C for up to 28 days. To examine hyphal induction in such conditions, yeast cultures were microscopically inspected at hourly intervals from 2-8h with a final reading at 24h. Fig 1 shows results of filamentation at a 5 mM final concentration of GlcNAc; however we note that all concentrations of GlcNAc tested produced similar levels of filamentation. Filamentation was further assayed using YEPD agar plus 10% fetal bovine serum (Sigma-aldrich, Milan, Italy) and Spider medium (nutrient broth 20 g/L, mannitol 20 g/L, K2HPO4 4 g/L, Bacto agar 27 g/L; pH 7.2) [23]. On these plates, 3 μl of standardized fungal suspension that contained 104, 103, 102 and 10 cells/μl, respectively, were spotted and plates were incubated, in duplicate, at 30 and 37°C for 7 days. Standard YEPD agar plates without serum were inoculated and used as a control.
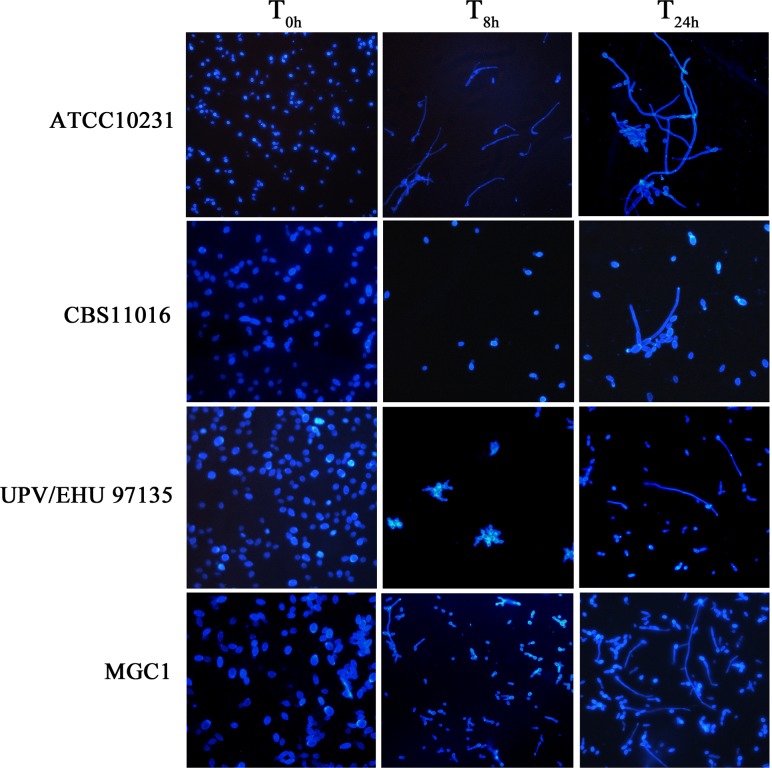
Fig 1. Morphologies of C. albicans and C. africana in YNB medium plus GlcNAc (5 mM final concentration).
Images were taken at different intervals of time (T0h, T8h and T24h). Results from different concentrations of GlcNAc (ranging from 0.195–100 mM) showed similar levels of filamentation (not shown).
100 μl of the standardized fungal suspensions (~107 cells/ml) were also used to inoculate 5 ml of YNB supplemented with 50 mM of galactose and 10 mM of GlcNAc in order to assess if our C. africana strains could be considered as NAG1 and/or DAC1 mutants according to previous studies [24].
Molecular tests
PCR primer design, amplification and sequencing of the NGT1 gene and NAG regulon
All PCR primers used to amplify and sequence the entire open reading frame (ORF) of the NGT1, HXK1, DAC1 and NAG1 genes are listed in Table 1.
Table 1. Primers used for amplification, sequencing and expression analysis.
| Primer name | Sequence (5'→3') | ORFsa | Amplicon size | Reference |
|---|---|---|---|---|
| NGT1-F | TACGGCATTGGAACAACCC | 19.5392 | 1804 bp | This study |
| NGT1-R | CCAACAGGATAGATTAGCCAACC | 19.5392 | -- | -- |
| NGT1-2Fb | ATGATTTGTGGATCAGTATTGGC | 19.5392 | 968 bp | This study |
| NGT1-2Rc | CGTCTTTCCAGTAAGGGTGAGT | 19.5392 | 942 bp | This study |
| NAG1-F | GCATTCGGTTGTTCTGGGAG | 19.2156 | 1098 bp | This study |
| NAG1-R | CCTTCAACTCTTATCTGTCGCC | 19.2156 | -- | -- |
| HXK1-Fa | CGCCAGTTGAATAAGATGGC | 19.2154 | 1178 bp | This study |
| HXK1-Ra | GGCGACAGATAAGAGTTGAAGG | 19.2154 | -- | -- |
| HXK1-Fbd | CGGTGTATTCTGCTGGGTG | 19.2154 | 898 bp | This study |
| HXK1-Rbe | CACCCAGCAGAATACACCG | 19.2154 | 896 bp | This study |
| DAC1-Fa | CTCCCAGAACAACCGAATGC | 19.2157 | 1459 bp | This study |
| DAC1-Ra | CGACGAATGGGCAACCTC | 19.2157 | -- | -- |
| DAC1-Fbf | TGCCACCCAGTCGAAACG | 19.2157 | 930 bp | This study |
| NGT1-RT-Rg | TGGACCGATATAAGCACCATC | 19.5392 | 491 bp | This study |
| NAG1-RT-F | GCATTGAGTGTTGGTATTTCCACC | 19.2156 | 195 bp | This study |
| NAG1-RT-R | ACTTTAATCCAGCGGCAGC | 19.2156 | -- | -- |
| HXK1-RT-F | CAAATTGTTGGGCAGTGACG | 19.2154 | 197 bp | This study |
| HXK1-RT-R | TGATAGGCAGCACCTATGGC | 19.2154 | -- | -- |
| DAC1-RT-F | CGTAACCTAGTCAAGTGGTCGC | 19.2157 | 198 bp | This study |
| DAC1-RT-R | TATCTGACGACTGGACTTCACG | 19.2157 | -- | -- |
| ACT1-RT-F | TCCAGAAGCTTTGTTCAGACCAGC | 19.5007 | 170 bp | [24] |
| ACT1-RT-R | TGCATACGTTCAGCAATACCTGGG | 19.5007 | -- | -- |
aOpen reading frames (ORFs) were downloaded from www.candidagenome.org and used for primer design
bThis primer was used in combination with the reverse primer NGT1-R
cThis primer was used in combination with the forward primer NGT1-F
dThis primer was used in combination with the reverse primer HXK1-Ra
eThis primer was used in combination with the forward primer HXK1-Fa
fThis primer was used in combination with the reverse primer DAC1-Ra
gThis primer was used in combination with the forward primer NGT1-2F.
Primers used for expression analysis are labeled with "RT".
Primers were designed with the Vector NTI program (version 10.3.0; Invitrogen, Italy) using the sequenced C. albicans genome (version A22-s04-m01-r03; www.candidagenome.org). Validation of primer specificity was performed using the web-based primer-BLAST tool (www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi).
For each gene, its ORF (Table 1) plus 1 kb of upstream and downstream sequence, was downloaded in FASTA format from the Candida genome database (www.candidagenome.org). A primer pair spanning ~200 bp or less upstream and downstream of each ORF was designed to amplify and sequence the entire coding region of each gene and additional internal sequencing primers were designed in cases where PCR primers did not suffice to cover the whole gene region in both directions (Table 1).
Total genomic DNA was extracted using a high-speed cell disruption method followed by phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation as described in Müller et al. [25].
In vitro amplifications were carried out in 50 μl volumes using DreamTaqTM PCR master mix (Fermentas, Milan, Italy) plus 1 μl of genomic DNA template and 0.5 μM of each primer pair (Table 1). All PCRs were performed in a MyCycler thermal cycler (Bio-Rad, Milan, Italy) with initial denaturation step at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 45 s, annealing at 52°C for 40 s, extension at 72°C for 90 s, and a final extension step of 10 min at 72°C.
A 3 μl aliquot of each PCR reaction was subjected to agarose gel electrophoresis to confirm the presence of amplified products, expected size and the absence of nonspecific fragments. The remaining amount of each amplicon was purified using the QIAquik PCR purification kit (Qiagen, Milan, Italy) and both strands were sequenced by MWG-Eurofins, Ebersberg, Germany (www.eurofinsgenomics.eu).
Electropherograms were inspected for errors and heterozygous mutations using FinchTV 1.4 software (www.geospiza.com) and sequence data were assembled using ContigExpress as implemented in Vector NTI.
Each DNA sequence was aligned using MEGA5 software [26] and compared with the corresponding wild-type ORF sequence (Table 1). Each nucleotide substitution leading to an amino acid change was inspected visually and recorded. Silent mutations were not considered.
All nucleotide sequences obtained in this study have been deposited in the GenBank database under the following accession numbers: KP052779, KP052780, KP165328, KP165330, KP165331, KP193952, KP193954, KP193955, KP193956, KP193958 and KP193959.
RNA extraction and RT-PCR assays
For gene expression studies, all isolates were grown in YNB with 2% galactose for 48h at 30°C. After incubation, cells were centrifuged at 1,455 g for 15 min, washed twice with sterile water and resuspended in YNB containing 5 mM GlcNAc. Yeast isolates were incubated at 30°C and RNA extraction was performed at 2.5, 8 and 24 hours. Total RNA was also obtained from fungal cultures grown in 2% galactose without GlcNAc induction.
RNA extraction from Candida spp. cultures was obtained by EuroGold Trifast reagent (EuroClone, Milan, Italy) following the manufacturer’s instructions with minor modifications. In brief, yeast cells were harvested by centrifugation at 1,455 g for 20 min, the pellet was resuspended in 1 ml of reagent and cells were lysed by mixing with an equal volume of acidified RNase-free glass beads (Sigma-Aldrich, Milan, Italy). One microgram of total RNA was subjected to DNaseI digestion following the manufacturer’s instructions (Sigma-Aldrich, Milan, Italy) and was reversely-transcribed using BioScipt M-MLV-Retro Transcriptase (Bioline, London, UK) with oligodT primer (EuroClone, Milan, Italy) at 42°C for 60 min followed by a denaturation step of 15 min at 70°C. Single strand cDNA was used in the PCR reaction to amplify NGT1, HXK1, DAC1, NAG1 and ACT1 (actin; housekeeping control) genes using BioTaq DNA polymerase (Bioline, London, UK). The primer sets used for expression analysis are listed in Table 1. Before RT-PCR, pilot experiments were performed to establish cycle number for the final analysis [27] and template dilutions to ensure that primer efficiency was as close as possible to 100% for all primer pairs. PCR conditions adopted were: 94°C 30 sec, 52°C 60 sec, 72°C 80 sec, 30 cycles and a final extension step of 10 min at 72°C.
The PCR products, after separation on an agarose gel, were acquired using a Kodak digital science 1D image analysis program (Eastman Kodak, Rochester, NY, USA). Densitometric analysis was performed using ImageJ software (http://imagej.nih.gov/ij). The expression level of all genes examined was normalized against the ACT1 gene, which was used as an internal housekeeping control gene. All experiments were repeated in triplicate. Statistical analysis of the data was performed by analysis of variance (One-Way-ANOVA) using Bonferroni’s multiple comparison post-test. A P-value <0.05 was considered significant.
Replacement of the C. africana mutated HXK1 gene by direct transformation with a wild-type HXK1 fragment from C. albicans
The DNA region carrying the mutation in the C. africana CBS11016 HXK1 gene was replaced using a 687 bp HXK1-fragment obtained from the C. albicans reference strain ATCC10231. The fragment was amplified in vitro using the following primer set: HXK1tr-fw GACTAGCATTAGTGGGTTGCG and HXK1-Rb CACCCAGCAGAATACACCG and the purified fragment was used to transform C. africana reference strain CBS11016 using the standard lithium acetate transformation method described previously [28] with the following modifications: transformants were recovered for 8h shaking in liquid YEPD media and then plated on SC media with 2% GlcNAc as the sole carbon source. Transformant colonies that were able to grow on the GlcNAc plates (after 5 days of growth) were selected and verified by sequencing. The MGC1 strain is a resulting verified transformant of the C. africana parent containing the wild-type C. albicans HXK1 fragment.
Results
Phenotypic tests
With the exception of the C. albicans control strain ATCC10231, C. africana isolates UPV/EHU 97135 and CBS11016 did not grow in tubes in the presence of different concentrations of GlcNAc after 28 days of incubation at 30°C. However, all fungal isolates were able to grow in liquid medium containing both galactose and GlcNAc after 24h of incubation.
Morphologically, considerable differences were observed between C. albicans and C. africana. After 8h of incubation in different GlcNAc concentrations (ranging from 0.195–100 mM), C. africana strains showed mostly yeast-like forms and a small number of germ tubes and pseudohyphae (~30% of the cells) after 24h (Fig 1; showing representative images in 5 mM GlcNAc). We note that the Spanish isolate UPV/EHU 97135 showed yeast cells arranged in irregular grape-like clusters (Fig 1).
C. albicans ATCC10231 formed germ-tubes efficiently after 2.5h of incubation in GlcNAc and after 8h the culture consisted nearly exclusively of long and short pseudohyphae (Fig 1).
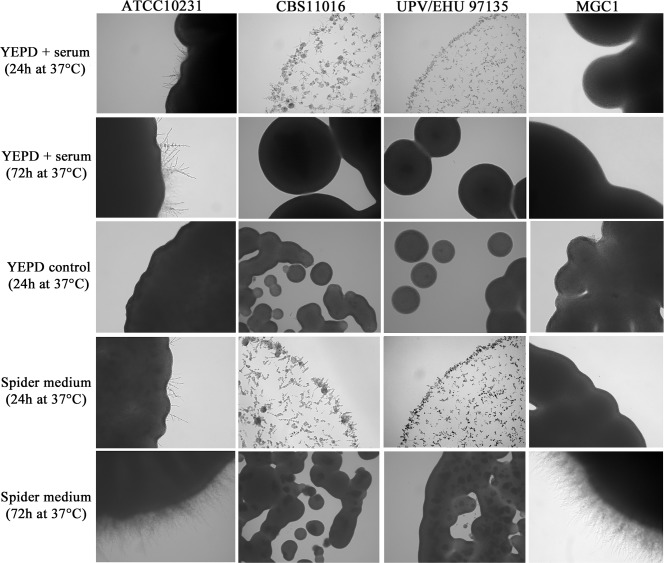
The degree of filamentation was also analyzed in specific solid filament-inducing media and the data revealed that after 24h of incubation at 30°C and 37°C on Spider medium and YEPD plus 10% serum, the C. albicans reference strain began to form hyphae (Fig 2).
Fig 2. Microscopic characteristics of C. albicans and C. africana isolates grown on YEPD plus 10% serum and Spider medium at 37°C for 24 and 72h.
YEPD control is also shown at 24h (at 72h the YEPD control was nearly identical to the 24h time point and thus is not shown). Images were taken from the first dilution (inoculum size ~104 cells/μl). Magnification 20x.
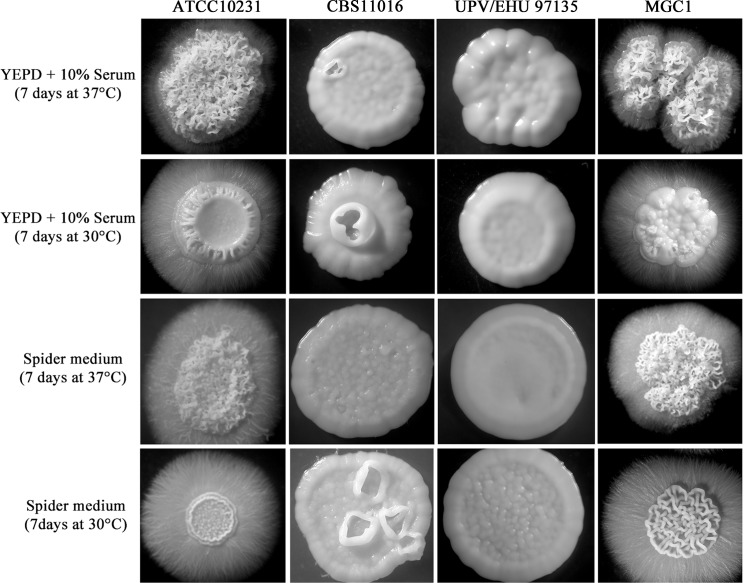
Upon microscopic examination, the colonies were composed mainly of pseudohyphal elements and after 7 days of incubation they were completely surrounded by hyphal outgrowth (Fig 3).
Fig 3. Morphologies of C. albicans and C. africana colonies obtained in this study after 7 days of incubation at 30°C and 37°C on YEPD plus 10% serum and Spider medium.
On the same agar media, C. africana isolates also showed increased filamentation after 24h at 30°C and 37°C, but their growth was delayed compared to C. albicans (Fig 2). The cultures were composed of hyphal forms (predominantly pseudohyphae) and budding yeast cells. On standard YEPD agar plates, used as a control, the colonies of C. africana strains were all smooth (Fig 2). After 48h of incubation on YEPD plus serum, C. africana isolates were mainly in the yeast form and their colonies appeared smooth, while in Spider medium, hyphae were abundant. After 72h at 30°C and 37°C, the colonies grown on YEPD plus serum were completely smooth, and in Spider medium, there was a notable increase of the yeast form. In contrast to C. albicans, after 7 days of incubation on these filament-inducing media, C. africana isolates showed only smooth colonies (Fig 3). Nevertheless, a high level of filamentation was observed in water plus 10% fetal bovine serum (vol/vol) at 30°C up to 7 days. In these conditions, C. africana produced filaments after 1h and no evident conversion back to the yeast form was observed during that time.
Genetic and expression analyses
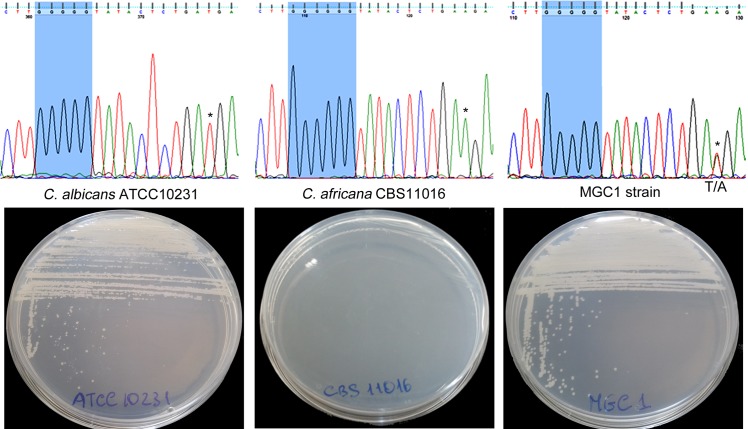
DNA sequence analysis of C. africana NGT1, HXK1, NAG1 and DAC1 genes revealed a number of characteristic nucleotide substitutions including a unique and distinctive nucleotide (guanine) insertion (Fig 4) that generated a stop codon (TGA) 154 bp downstream of the ATG start codon of the HXK1 gene. This mutation was also found in other C. africana strains from different geographical origins (ten Chinese strains and one German strain) [3, 21] included in this study. On the contrary, the C. albicans ATCC10231 strain lacked this polymorphism (Fig 4).
Fig 4. Partial HXK1-gene sequencing of C. albicans ATCC10231, C. africana CBS11016 and MGC1 strain showing the region containing the guanine insertion.
In the MGC1 strain, a heterozygous site containing both C. albicans and C. africana specific nucleotides is also shown (* indicates the C. albicans and C. africana peaks that overlap in the heterozygous site). The respective growth of the strains in YNB medium plus GlcNAc is also shown.
To verify whether the guanine (G) insertion was directly responsible for the inability to assimilate GlcNAc by C. africana, the DNA region containing this mutation was replaced in the CBS11016 C. africana strain using a specific 687 bp HXK1-fragment derived from the C. albicans reference strain. Partial sequencing of the HXK1 gene of the resulting transformant (designated MGC1) confirmed the absence of the distinctive G insertion and the presence of both C. albicans and C. africana alleles as demonstrated by heterozygous sites (Fig 4).
Phenotypically, the MGC1 strain was able to grow in media containing GlcNAc as the sole carbon source (Fig 4) and its colonies resembled those of the C. albicans ATCC10231 strain when grown under filament-inducing conditions (Fig 3).
Comparison of DNA sequencing results of the NGT1 gene and the orf19.5392 C. albicans reference sequence showed a complete coding sequence with a single non-synonymous substitution (1010A→G), which replaces lysine (K) with arginine (R) within the Ngt1 protein. This neutral substitution was the only mutation detected in the NGT1 gene of our C. africana isolates.
Sequence alignment of the DAC1 genes revealed several nucleotide substitutions, most of which were silent or neutral mutations in the respective proteins. Only one, the 640A→G substitution, caused the replacement of an aspartic acid (D) with asparagine (N) within the C. africana Dac1 protein.
Regarding the NAG1 gene, although several base substitutions were detected, two in particular, located at nt 334 and 443, caused the amino acidic substitution Glu→Lys and Ala→Thr at position 112 and 145, respectively, in all C. africana Nag1 proteins. Nevertheless, the putative 3D structure, predicted by comparative homology modeling (http://toolkit.tuebingen.mpg.de/hhpred) using the Escherichia coli glucosamine 6-phosphate deaminase protein (PDB: 1HOR) as a template, revealed no structural differences (data not shown).
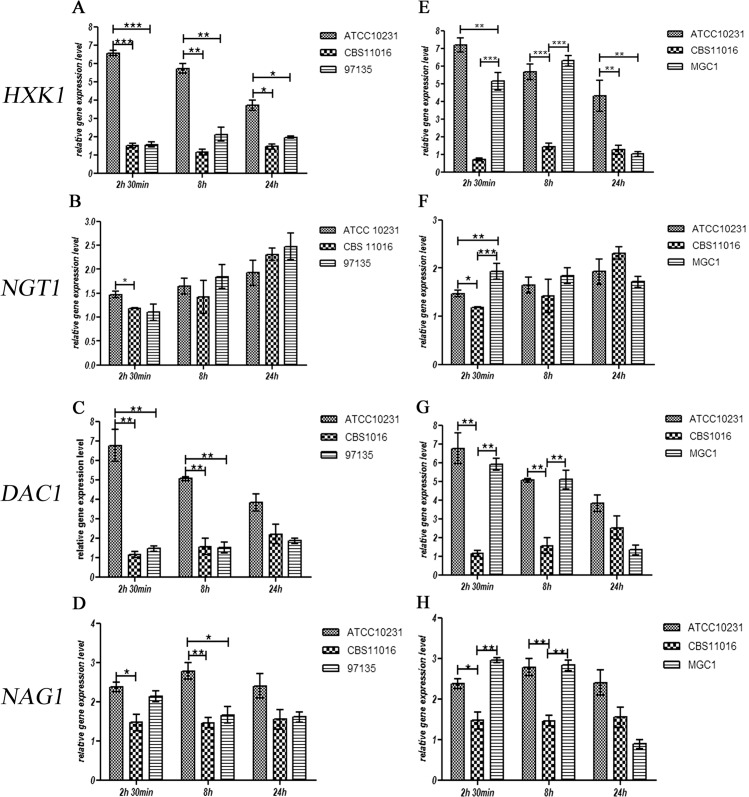
The transcription of all genes involved in GlcNAc catabolism was also examined indicating that, in C. albicans ATCC10231, the presence of GlcNAc in the medium was able to evoke a strong initial induction of expression of the HXK1 gene, which decreased progressively over time (Fig 5A).
Fig 5. Effect of GlcNAc on mRNA expression levels of the HXK1, NGT1, DAC1 and NAG1 genes in C. albicans ATCC10231, C. africana CBS1016, C. africana UPV/EHU 97135 (A-D) and the MGC1 strain (E-H).
Values are normalized to ACT1 as an internal control. Data are expressed as fold changes with respect to untreated cells. The experiments were performed in triplicate and results are expressed as means ± SEM. Statistical analysis were performed using a One-Way-ANOVA with Bonferroni’s Multiple comparison post-test. A P value < 0.05 was considered significant.
Different results were obtained from C. africana strains in which the level of HXK1 gene expression was much lower, compared with data obtained from C. albicans strain (P-value <0.05) (Fig 5A). These results suggest that the presence of a premature stop codon in the C. africana HXK1 mRNA leads to an instable transcript that is rapidly degraded.
Previous studies showed that in C. albicans HXK1 null mutants, the expression levels of the others genes involved in GlcNAc catabolism are altered [24, 29]. To better understand the effect of the naturally occurring C. africana non-functional HXK1 gene, the expression levels of NGT1, DAC1, and NAG1 genes were also investigated. The transcription of the GlcNAc transporter, in all Candida strains, does not seem to be impaired by the integrity of the HXK1 gene. As shown in Fig 5B, the comparative analysis among Candida strains showed no significant differences between NGT1 mRNA levels.
The effect on DAC1 and NAG1 transcriptional rate was significantly different; in fact, in C. africana strains, the transcriptional level of the DAC1 gene was much lower at 2.5h and 8h compared to C. albicans (Fig 5C). The effect exerted on NAG1 expression was less pronounced (although statistically significant), and very similar to that observed for the DAC1 gene expression (Fig 5D).
In an attempt to clarify whether the differences observed at the transcriptional level for these selected genes were linked to a non-functional HXK1 gene, the mutated allele in C. africana CBS11016 strain was replaced by homologous recombination using a functional HXK1 gene derived from C. albicans. The resulting transformant (strain MGC1) displayed a full restoration of the HXK1 gene expression that was very similar to that observed for the C. albicans wild-type strain (Fig 5E). However the HXK1 expression levels decreased drastically at 24h, reaching a level similar to that observed for C. africana CBS11016 (Fig 5E). For the NGT1 gene, results showed a possible independence of its transcription from the HXK1 gene. In fact it is evident that the restoration of the functional HXK1 allele in C. africana had no influence on the GlcNAC transporter NGT1 in this timeframe (Fig 5F). On the contrary, the transcription of the DAC1 and NAG1 genes in C. africana appears to be considerably influenced by HXK1 restoration; in fact, comparison of DAC1 mRNA levels between the C. africana CBS11016 strain and the MGC1 strain highlights a strong effect exerted by a functional HXK1, producing a level of DAC1 transcripts that is very similar to that of the C. albicans wild-type strain. Nevertheless, no differences were observed among the three Candida strains at 24h (Fig 5G).
Similar results were also obtained for NAG1 gene expression (Fig 5H).
Discussions
In last few years, many epidemiological studies reporting the clinical isolation of C. africana have appeared in the scientific literature and the data obtained revealed that this fungus is particularly adapted to colonize human genitalia and infects mainly such organs [9, 17, 19, 21, 30, 31, 32, 33, 34].
The propensity to cause genital infections may be the result of its unique and distinctive genetic background [7] which makes this fungus particularly interesting to study [1]. In fact, unlike typical C. albicans isolates, C. africana isolates display a reduced virulence, possess a limited pathogenic potential [17, 35] and display a number of phenotypic defects including the inability to metabolize several organic compounds [3]. Therefore, the existence of this natural mutant within the C. albicans population [10] offers a powerful model for understanding the molecular basis of pathogenicity, phenotypic diversification and adaptation to mammalian hosts for pathogenic members of the Candida clade.
Through this study, we were able to demonstrate that the lack of GlcNAc utilization by C. africana is due to a single nucleotide (G residue) insertion resulting in a frameshift mutation that, created a premature stop codon within the HXK1-gene encoding GlcNAc kinase. Thus C. africana can be considered a natural HXK1 mutant as also evidenced by positive growth of our isolates in medium containing galactose plus GlcNAc, which impairs the development of the strains lacking the DAC1 and/or NAG1 genes, but not of the HXK1 mutants [14, 24]. In addition, the replacement of the mutated HXK1 allele using a functional allele from C. albicans, restored the gene function and GlcNAc assimilation in C. africana (Fig 4)
Previous studies have already genetically well-characterized the GlcNAc catabolic pathway in C. albicans [24, 29, 36] and revealed an important role of the NAG regulon in the biology and virulence of this fungus, especially regarding the function of the HXK1 gene, which seems to be involved in various cellular processes in this species [29].
Disruption of the entire NAG cluster in C. albicans revealed some interesting phenotypic characteristics including the ability of the triple mutant to hyperfilament under stress-induced conditions [36]. This intrinsic morphological property has also been reported for HXK1 null mutants [29, 37] and further confirmed in this study for C. africana.
Rao et al. [29] reported that the homozygous HXK1 mutant, after 2 days of incubation at 30°C on YEPD plus serum, started to form wrinkled colonies which were completely surrounded by hyphae after 6 days in such conditions. Similar results were also obtained in a separate study [37] and are, in part, consistent with our data. In fact we observed that C. africana strains showed a strong increase in hyphal formation on solid media during the first 24h of incubation but their colonies soon turned smooth and no filamentation was observed after 7 days of incubation (Fig 3). However C. africana was able to filament abundantly in 10% serum in water; a similar behavior was also previously observed in Candida dubliniensis, a species phylogenetically closely related to C. albicans that fails to form hyphae in YEPD supplemented with 10% serum but not in nutrient-poor media [38]. Therefore, the strong tendency of C. africana to hyperfilament rapidly and quickly revert to the yeast-phase in specific rich agar media raises the hypothesis that nutrient-sensing mechanisms in this biovariant could be differentially regulated compared with C. albicans and may partly explain its low degree of virulence and pathogenicity observed in recent studies [17, 35].
In our study, differences were also seen in gene expression profiles. In fact although the expression levels of the HXK1 gene were lower in all C. africana isolates (Fig 5A), considerable differences were observed regarding other genes involved in GlcNAc metabolism except NGT1 whose expression, in agreement with previous studies, does not appear to be controlled by HXK1 [24, 39]. Our data (Fig 5) highlight a strong influence of a functional GlcNAc kinase in the transcription of both DAC1 and NAG1 genes. These results are in accord with a conceptual scheme presented by Rao et al. [29] in which the HXK1 gene plays a pivotal role in different cellular processes including DAC1 and NAG1 expression, and partially in accord with other published data [24] in which a C. albicans HXK1 null mutant showed a reduced, but consistent transcriptional level for these genes, especially for the NAG1 gene. Overall these data confirm that the Hxk1 enzyme could play additional roles other than its traditional metabolic role as a kinase [29].
We believe that the existence of a group of strains, referred as C. africana, within C. albicans populations, with distinct properties such as reduced virulence and pathogenicity, suggests the possibility for the presence of other novel biological functions, not only for the HXK1 gene, but also for other aspects of the biology and genetics of C. albicans. Therefore C. africana could represent a useful natural model for future comparative genomics and transcriptomics studies in order to better understand the molecular mechanisms that make C. albicans the most predominant fungal pathogen for humans.
Acknowledgments
The authors wish to thank Dr. Guilliermo Quindos (Universidad del País Vasco, Bilbao, Spain) for providing the C. africana UPV/EHU 97135 strain.
We also thank Dr. Hans-Jürgen Tietz (Institut für Pilzkrankheiten, Berlin, Germany) and Dr. Shangrong Fan (Peking University Shenzhen Hospital, Shenzhen, China) for supplying the German and Chinese C. africana isolates used to confirm the HXK1 gene mutation reported in this study.
We are also very grateful to Dr. Aaron Hernday for helpful technical discussions.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Romeo O, Criseo G. Candida africana and its closest relatives. Mycoses. 2011;54: 475–486. 10.1111/j.1439-0507.2010.01939.x [DOI] [PubMed] [Google Scholar]
- 2.Romeo O, Tietz HJ, Criseo G. Candida africana: is it a fungal pathogen? Curr Fungal Infect Rep. 2013;7: 192–197. [Google Scholar]
- 3.Tietz HJ, Hopp M, Schmalreck A, Sterry W, Czaika V. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses. 2001;44: 437–445. [DOI] [PubMed] [Google Scholar]
- 4.Criseo G, Scordino F, Romeo O. Current methods for identifying clinically important cryptic Candida species. J Microbiol Methods. 2015;111: 50–56. 10.1016/j.mimet.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 5.Forche A, Schönian G, Gräser Y, Vilgalys R, Mitchell TG. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet Biol. 1999;28: 107–125. [DOI] [PubMed] [Google Scholar]
- 6.Ciardo DE, Schär G, Böttger EC, Altwegg M, Bosshard PP. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J Clin Microbiol. 2006;44: 77–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, et al. Molecular phylogenetics of Candida albicans. Eukaryot Cell. 2007;6: 1041–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romeo O, Criseo G. Morphological, biochemical and molecular characterisation of the first Italian Candida africana isolate. Mycoses. 2009;52: 454–457. 10.1111/j.1439-0507.2008.01630.x [DOI] [PubMed] [Google Scholar]
- 9.Ngouana TK, Krasteva D, Drakulovski P, Toghueo RK, Kouanfack C, Ambe A, et al. Investigation of minor species Candida africana, Candida stellatoidea and Candida dubliniensis in the Candida albicans complex among Yaoundé (Cameroon) HIV-infected patients. Mycoses. 2015;58: 33–9. 10.1111/myc.12266 [DOI] [PubMed] [Google Scholar]
- 10.Odds FC. Molecular phylogenetics and epidemiology of Candida albicans. Future Microbiol. 2010;5: 67–79. 10.2217/fmb.09.113 [DOI] [PubMed] [Google Scholar]
- 11.Naseem S, Parrino SM, Buenten DM, Konopka JB. Novel roles for GlcNAc in cell signaling. Commun Integr Biol. 2012;5: 156–159. 10.4161/cib.19034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tietz HJ, Küssner A, Thanos M, Pinto De Andreade M, Presber W, et al. Phenotypic and genotypic characterization of unusual vaginal isolates of Candida albicans from Africa. J Clin Microbiol. 1995;33: 2462–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang G, Yi S, Sahni N, Daniels KJ, Srikantha T, Soll DR. N-acetylglucosamine induces white to opaque switching, a mating prerequisite in Candida albicans. PLoS Pathog. 2010;6: e1000806 10.1371/journal.ppat.1000806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konopka JB. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica (Cairo). 2012; 10.6064/2012/489208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang G. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence. 2012;3: 251–61. 10.4161/viru.20010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez FJ, Konopka JB. Identification of an N-acetylglucosamine transporter that mediates hyphal induction in Candida albicans. Mol Biol Cell. 2007;18: 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borman AM, Szekely A, Linton CJ, Palmer MD, Brown P, Johnson EM. Epidemiology, antifungal susceptibility, and pathogenicity of Candida africana isolates from the United Kingdom. J Clin Microbiol. 2013;51: 967–972. 10.1128/JCM.02816-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romeo O, De Leo F, Criseo G. Adherence ability of Candida africana: a comparative study with Candida albicans and Candida dubliniensis. Mycoses. 2011;54: 57–61. [DOI] [PubMed] [Google Scholar]
- 19.Alonso-Vargas R, Elorduy L, Eraso E, Cano FJ, Guarro J, Pontón J, et al. Isolation of Candida africana, probable atypical strains of Candida albicans, from a patient with vaginitis. Med Mycol. 2008;46: 167–170. [DOI] [PubMed] [Google Scholar]
- 20.Romeo O, Criseo G. First molecular method for discriminating between Candida africana, Candida albicans, and Candida dubliniensis by using hwp1 gene. Diagn Microbiol Infect Dis. 2008;62: 230–233. 10.1016/j.diagmicrobio.2008.05.014 [DOI] [PubMed] [Google Scholar]
- 21.Shan Y, Fan S, Liu X, Li J. Prevalence of Candida albicans-closely related yeasts, Candida africana and Candida dubliniensis, in vulvovaginal candidiasis. Med Mycol. 2014;52: 636–40. 10.1093/mmy/myu003 [DOI] [PubMed] [Google Scholar]
- 22.Kurtzman CP, Fell JW. The yeasts, a taxonomy study, 4th ed. Amsterdam: Elsevier Science; 1998. [Google Scholar]
- 23.Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009;5: e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naseem S, Gunasekera A, Araya E, Konopka JB. N-acetylglucosamine (GlcNAc) induction of hyphal morphogenesis and transcriptional responses in Candida albicans are not dependent on its metabolism. J Biol Chem. 2011;286: 28671–28680. 10.1074/jbc.M111.249854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller FM, Werner KE, Kasai M, Francescani A, Chanock SJ, Walsh TJ. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J Clin Microbiol. 1998;36: 1625–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prediger EA. Quantitating mRNAs with relative and competitive RT-PCR. Methods Mol Biol. 2001;160: 49–63. [DOI] [PubMed] [Google Scholar]
- 28.Nobile CJ, Mitchell AP. Large-scale gene disruption using the UAU1 cassette. Methods Mol Biol. 2009;499: 175–194. 10.1007/978-1-60327-151-6_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rao KH, Ghosh S, Natarajan K, Datta A. N-acetylglucosamine kinase, HXK1 is involved in morphogenetic transition and metabolic gene expression in Candida albicans. PLoS One. 2013;8: e53638 10.1371/journal.pone.0053638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romeo O, Criseo G. Molecular epidemiology of Candida albicans and its closely related yeasts Candida dubliniensis and Candida africana. J Clin Microbiol. 2009;47: 212–214. 10.1128/JCM.01540-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieng Y, Sow D, Ndiaye M, Guichet E, Faye B, Tine R, et al. Identification of three Candida africana strains in Senegal. J Mycol Med. 2012;22: 335–340. 10.1016/j.mycmed.2012.07.052 [DOI] [PubMed] [Google Scholar]
- 32.Nnadi NE, Ayanbimpe GM, Scordino F, Okolo MO, Enweani IB, Criseo G, et al. Isolation and molecular characterization of Candida africana from Jos, Nigeria. Med Mycol. 2012;50: 765–767. 10.3109/13693786.2012.662598 [DOI] [PubMed] [Google Scholar]
- 33.Sharma C, Muralidhar S, Xu J, Meis JF, Chowdhary A. Multilocus sequence typing of Candida africana from patients with vulvovaginal candidiasis in New Delhi, India. Mycoses. 2014;57: 544–552. 10.1111/myc.12193 [DOI] [PubMed] [Google Scholar]
- 34.Yazdanparast SA, Khodavaisy S, Fakhim H, Shokohi T, Haghani I, Nabili M, et al. Molecular characterization of highly susceptible Candida africana from vulvovaginal candidiasis. Mycopathologia. 2015; 10.1007/s11046-015-9924-z [DOI] [PubMed] [Google Scholar]
- 35.Pagniez F, Jimenez-Gil P, Mancia A, Le Pape P. Étude comparative in vivo de la virulence de Candida africana et de C. albicans stricto sensu. J Mycol Med. 2015;25: e107. [Google Scholar]
- 36.Singh P, Ghosh S, Datta A. Attenuation of virulence and changes in morphology in Candida albicans by disruption of the N-acetylglucosamine catabolic pathway. Infect Immun. 2001;12: 7898–7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wendland J, Hellwig D, Walther A, Sickinger S, Shadkchan Y, Martin R, et al. Use of the Porcine Intestinal Epithelium (PIE)-Assay to analyze early stages of colonization by the human fungal pathogen Candida albicans. J Basic Microbiol. 2006;46: 513–523. [DOI] [PubMed] [Google Scholar]
- 38.O'Connor L, Caplice N, Coleman DC, Sullivan DJ, Moran GP. Differential filamentation of Candida albicans and Candida dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryot Cell. 2010;9: 1383–1397. 10.1128/EC.00042-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao KH, Ruhela D, Ghosh S, Abdin MZ, Datta A. N-acetylglucosamine kinase, HXK1 contributes to white-opaque morphological transition in Candida albicans. Biochem Biophys Res Commun. 2014;445: 138–144. 10.1016/j.bbrc.2014.01.123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.