Abstract
Centrosome amplification is a common feature of both solid and hematological human malignancies. Extra centrosomes are not merely innocent bystanders in cancer cells, but rather promote tumor progression by disrupting normal cellular architecture and generating chromosome instability. Consequently, centrosome amplification correlates with advanced tumor grade and overall poor clinical prognosis. By contrast, extra centrosomes are adversely tolerated in non-transformed cells and hinder cell proliferation. This suggests that in addition to acquiring extra centrosomes, cancer cells must also adapt to overcome the deleterious consequences associated with them. Here, we review evidence that implicates core components of the Hippo tumor suppressor pathway as having key roles in both the direct and indirect regulation of centrosome number. Intriguingly, functional inactivation of the Hippo pathway, which is common across broad spectrum of human cancers, likely represents one key adaptation that enables cancer cells to tolerate extra centrosomes.
Keywords: YAP, TAZ, LATS, p53, tetraploid, MST
Introduction
The centrosome is the major microtubule nucleating structure in mammalian cells and plays a significant role in regulating diverse cellular functions, including cilia formation, cell signaling, cell migration, establishment of cell polarity, and mitosis (reviewed extensively in this issue and elsewhere) (Chavali et al., 2014, Doxsey et al., 2005, Godinho and Pellman, 2014, Nigg and Raff, 2009). Regulation of centrosome number is tightly controlled, as the presence of too many or too few centrosomes is known to significantly impair the proliferation and viability of non-transformed cells (Ganem et al., 2014, Ganem et al., 2009, Holland et al., 2012, Lambrus et al., 2015, Marthiens et al., 2013, Wong et al., 2015). However, somewhat paradoxically, centrosome amplification is prevalent in human tumors, suggesting that extra centrosomes may provide a fitness benefit to cancer cells (D'Assoro et al., 2002, Ganem et al., 2009, Ghadimi et al., 2000, Godinho and Pellman, 2014, Lingle et al., 2002, Sluder and Nordberg, 2004). Understanding the mechanisms that give rise to extra centrosomes, deciphering how cancer cells adapt to tolerate extra centrosomes, and determining the potential beneficial effects of extra centrosomes for cancer cells represent several key unresolved questions in cancer cell biology. Interestingly, several studies now implicate the Hippo tumor suppressor pathway, which is frequently inactivated in human tumors, as having key roles in both the direct and indirect regulation of centrosome number. Here, we provide a general overview of Hippo pathway signaling, introduce mechanisms that link centrosome amplification to activation of the Hippo pathway, and discuss the role of Hippo pathway core components in regulating centrosome number.
Causes and Consequences of Centrosome Amplification
Centrosome number, similar to chromosome number, is highly regulated within cells. Cells in G0 or G1 phase typically contain a single centrosome, comprised of two orthogonal centrioles surrounded by a cloud of pericentriolar material. Similar to DNA replication, centrosome duplication occurs only once during S-phase and generates two centrosomes for G2/M (Firat-Karalar and Stearns, 2014, Nigg and Stearns, 2011, Sluder, 2014, Tsou and Stearns, 2006a, Tsou and Stearns, 2006b). The two centrosomes present during mitosis facilitate the formation of a bipolar mitotic spindle and thus promote high fidelity chromosome segregation during anaphase (Hinchcliffe, 2014). Following cell division, each daughter cell inherits a single centrosome and the replication cycle repeats.
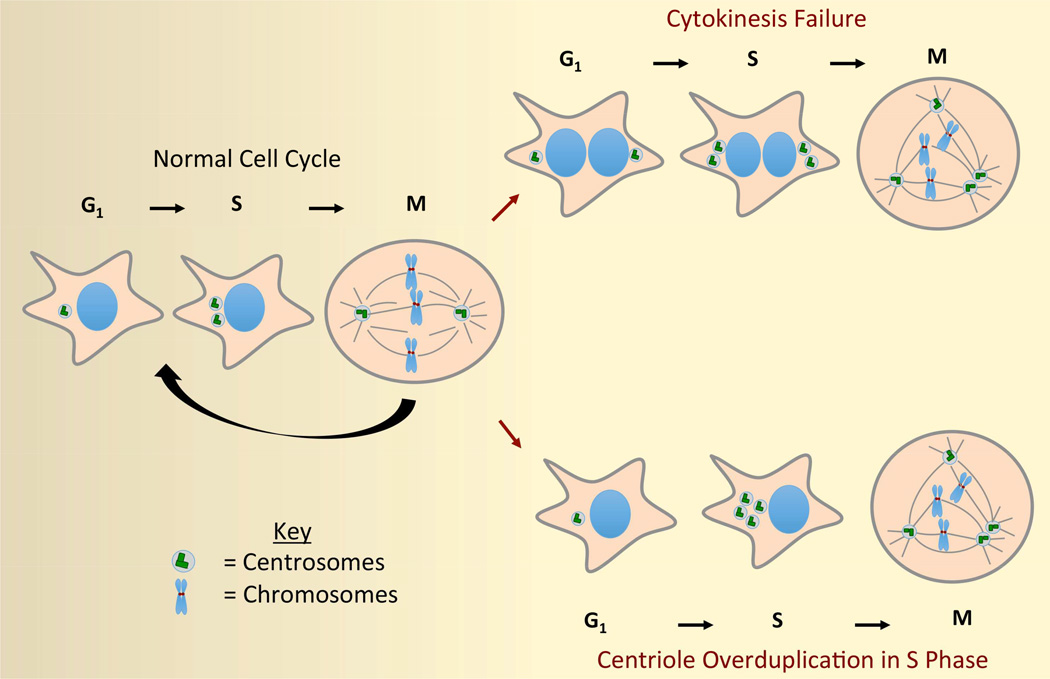
While maintenance of centrosome number is highly regulated in non-transformed cells, errors do occasionally arise that lead to centrosome amplification. Extra centrosomes primarily arise via two distinct, yet non-mutually exclusive, mechanisms (Figure 1). Extra centrosomes can be generated by mitotic or cytokinetic failures, which produce tetraploid cells with twice the normal number of centrosomes (Davoli and de Lange, 2011, Ganem et al., 2007, Meraldi et al., 2002). Alternatively, extra centrosomes can arise from cellular and/or genetic defects that promote centriole overduplication (Firat-Karalar and Stearns, 2014, Nigg and Stearns, 2011, Sluder, 2014).
Figure 1. Mechanisms of centrosome amplification.
Extra centrosomes predominantly arise from cytokinesis failure or centriole overduplication. Following cytokinesis failure, two centrosomes are present in G1 interphase cells. These centrosomes replicate during S-phase and the resulting 4 centrosomes promote multipolar cell division during the next mitosis. Abnormal centriole duplication arises during S-phase and can produce a variable number of centrosomes in the subsequent mitosis.
Regardless of the mechanisms underlying centrosome amplification, it is now well recognized that excess centrosomes are detrimental to the viability of non-transformed cells. Most notably, extra centrosomes have the capacity to greatly disrupt mitotic spindle formation and accurate chromosome segregation. Cells that enter mitosis with more than two centrosomes are predisposed to forming multipolar spindles (Brinkley, 2001, Nigg, 2002). Unless resolved prior to anaphase onset, multipolar spindles lead to catastrophic multipolar anaphase, the generation of grossly aneuploid daughter cells, and cell death (Ganem et al., 2009). This harmful effect of extra centrosomes can have profound consequences on organismal health. In mice, induced centrosome amplification in the brain promotes significant cell death through multipolar division, ultimately leading to microcephaly (Marthiens et al., 2013).
Extra centrosomes also appear to disrupt normal cell proliferation through mechanisms that are independent of abnormal mitosis and aneuploidy. Remarkably, it has been demonstrated that the presence of even a single extra centrosome in non-transformed cells is sufficient to activate the p53 tumor suppressor pathway and impede further cell proliferation (Ganem et al., 2014, Holland et al., 2012). This holds true irrespective of whether extra centrosomes are generated by cytokinesis failure or centriole overduplication. Unsurprisingly, non-transformed cells with extra centrosomes are selected against in long-term culture experiments (Chiba et al., 2000, Ganem et al., 2009, Godinho et al., 2014).
Cancer Cells Adapt to Tolerate Extra Centrosomes
While centrosome number is strictly regulated in non-transformed cells, the opposite is true for many tumor cells. It is firmly established that centrosome amplification is a hallmark of human cancers (D'Assoro et al., 2002, Ganem et al., 2009, Ghadimi et al., 2000, Godinho and Pellman, 2014, Lingle et al., 2002, Sluder and Nordberg, 2004). This raises an obvious paradox: If extra centrosomes are so poorly tolerated by non-transformed cells, then why are they so common in human malignancies? One possibility is that there are positive selective pressures that promote the accumulation of extra centrosomes in cancer cells. For example, extra centrosomes are known to promote chromosome instability (reviewed below) (Ganem et al., 2009, Silkworth et al., 2009). Chromosome instability (CIN) is defined by the persistently elevated rate of whole chromosome missegregation during cell division (Lengauer et al., 1997). CIN generates significant genetic heterogeneity within tumors and can enable the outgrowth of cells that have acquired growth advantages (Davoli et al., 2013, Pavelka et al., 2010, Rancati et al., 2008, Selmecki et al., 2006, Sheltzer et al., 2011, Sotillo et al., 2010, Thompson and Compton, 2010). Thus, acquisition of a CIN phenotype may be a major positive selective pressure for cancer cells to accumulate extra centrosomes. Extra centrosomes have also been shown to disrupt cell polarity, promote asymmetric cell division, alter cellular signaling, and promote tumor cell invasiveness, any or all of which may promote tumor growth and progression (Basto et al., 2008, Godinho et al., 2014, Mahjoub and Stearns, 2012).
Another reason why extra centrosomes are more common in cancer cells could be that they are simply better tolerated. It is now obvious that cancer cells acquire several characteristics that make them more permissive of the negative effects imparted by extra centrosomes. One clear adaptation made by cancer cells to cope with extra centrosomes is to reduce the propensity of cells with extra centrosomes to undergo chaotic multipolar cell divisions. To accomplish this, most cancer cells cluster excess centrosomes into two spindle poles during mitosis, enabling a relatively normal bipolar anaphase (Basto et al., 2008, Kwon et al., 2008). While this mechanism saves cells from catastrophic mitosis and likely cell death, it is not without consequences. Cancer cells with extra centrosomes pass through a transient ‘multipolar spindle intermediate’ prior to centrosome clustering, during which merotelic kinetochore-microtubule attachment errors accumulate (Ganem et al., 2009, Silkworth et al., 2009). Merotelic attachments (defined as single kinetochores attached to two spindle poles) are known to promote chromosome missegregation (Cimini et al., 2001, Cimini et al., 2003, Compton, 2011, Salmon et al., 2005, Thompson and Compton, 2008, Thompson and Compton, 2011). Thus, extra centrosomes, even if clustered into two poles to preserve cell viability, continue to promote whole chromosome segregation errors with resultant CIN.
Additional adaptive mechanisms made by cancer cells to tolerate extra centrosomes remain less well defined. For example, although extra centrosomes are known to stimulate the p53 pathway and limit the proliferation of non-transformed cells, the mechanisms underlying this response are poorly understood. Recent studies demonstrate that centrosome amplification activates the Hippo tumor suppressor pathway, which indirectly stabilizes p53 (Ganem et al., 2014). This suggests that Hippo pathway inactivation, which is a widespread feature of human cancers, may represent one common adaptation that enables cancer cells to better tolerate extra centrosomes. Below, we describe canonical Hippo pathway signaling and speculate as to how extra centrosomes may trigger activation of this pathway.
The Hippo Tumor Suppressor Pathway
The Hippo tumor suppressor pathway is a conserved regulator of cellular proliferation, differentiation, and death. Canonically, the major function of the Hippo pathway is to restrain organ size (Pan, 2010, Yu and Guan, 2013), as loss of Hippo pathway activity is well known to promote tissue overgrowth and tumor development (Camargo et al., 2007, Dong et al., 2007, Lu et al., 2010, Song et al., 2010, Zhou et al., 2009a). However, recent studies have demonstrated that the Hippo pathway is also regulated by complex inputs that monitor cell-cell adhesion, cell-matrix adhesion, G-protein coupled receptor (GPCR) signaling, and contractile tension from the actin cytoskeleton (Aragona et al., 2013, Dupont et al., 2011, Ganem et al., 2014, Halder et al., 2012, Paramasivam et al., 2011, Wada et al., 2011, Yu et al., 2012, Zhao et al., 2012, Zhao et al., 2007). Indeed, several conditions known to induce cell cycle arrest, including contact inhibition, loss of cell attachment, and mitotic failure are now known to activate the Hippo pathway (Aragona et al., 2013, Aylon et al., 2006, Ganem et al., 2014, Zhao et al., 2012, Zhao et al., 2007).
The core components of the Hippo pathway, which are conserved in mammals, were first identified in genetic screens for tumor suppressors in Drosophila melanogaster. These include Hippo (Mammalian sterile 20-like kinases 1 and 2 (MST1 and MST2) orthologs in mammals) (Harvey et al., 2003, Jia et al., 2003, Pantalacci et al., 2003, Udan et al., 2003, Wu et al., 2003), Warts (Large tumor suppressor kinases 1 and 2 (LATS1 and LATS2) orthologs in mammals) (Justice et al., 1995, Xu et al., 1995), Salvador (SAV1 ortholog in mammals) (Kango-Singh et al., 2002, Tapon et al., 2002), and Mats (Mps1 binder (MOB) ortholog in mammals) (Lai et al., 2005). Loss of function mutations in these genes lead to increased cell proliferation, reduced cell death, and increased organ size in flies (e.g. overgrown eyes and wings). Similarly, loss of Hippo pathway activity promotes tissue overgrowth and tumor development in mice (Camargo et al., 2007, Dong et al., 2007, Lu et al., 2010, Song et al., 2010, Zhou et al., 2009a). For simplicity, the mammalian gene/protein names will be used for the purposes of this review.
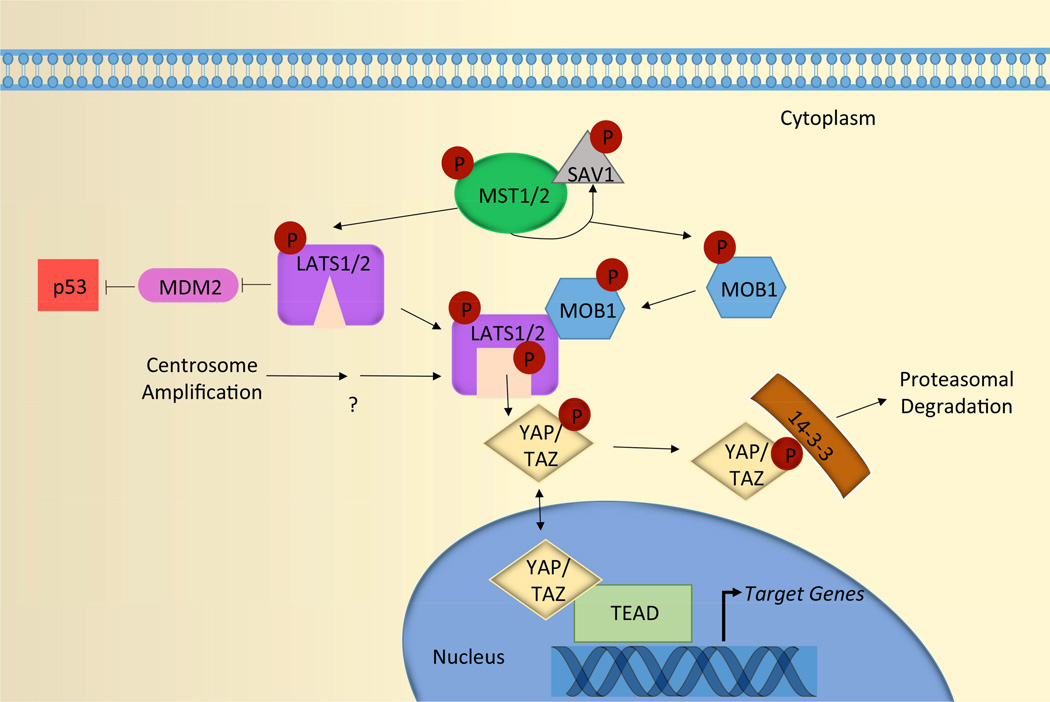
The main function of the Hippo pathway is to negatively regulate the oncogenic transcriptional co-activators yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ) (Pan, 2010, Yu and Guan, 2013). This regulation is primarily accomplished through activation of the kinases LATS1 and LATS2, which phosphorylate YAP and TAZ to promote their inactivation (Figure 2) (Yu and Guan, 2013, Zhao et al., 2010a). Phosphorylated YAP and TAZ bind to 14-3-3, which sequesters YAP/TAZ in the cytoplasm where they are subsequently proteasomally degraded (Hong and Guan, 2012). YAP/TAZ can also be sequestered at both tight and adherens junctions through direct binding to proteins that localize there (Avruch et al., 2012, Bertini et al., 2009, Zhao et al., 2010b, Zhao et al., 2011, Oka et al., 2008). Ultimately, activation of the Hippo pathway prevents YAP and TAZ from entering the nucleus and activating the transcriptional enhancer activation domain (TEAD)-family of transcription factors to initiate the expression of genes important for cell growth and survival (Pan, 2010, Yu and Guan, 2013, Zhao et al., 2010a, Zhao et al., 2010b, Zhao et al., 2011).
Figure 2. Canonical Hippo pathway signaling.
MST1/2 kinases, in complex with the scaffolding protein SAV1, directly phosphorylate LATS1/2 and MOB1. Phosphorylated MOB1 binds to the auto-inhibitory regions of LATS1/2, which releases LATS1/2 from their inhibitory state and enables auto-phosphorylation on their activation loops. Together, the coordinated actions of MST1/2 and MOB fully activate LATS1/2. Active LATS1/2 then phosphorylate both YAP and TAZ. Phosphorylated YAP and TAZ are sequestered in the cytoplasm through binding to 14-3-3, where they are subsequently proteasomally degraded. Consequently, activation of the Hippo pathway prevents YAP and TAZ from entering the nucleus and activating the TEAD-family of transcription factors to initiate the transcription of genes important for cell growth and survival. In addition, active LATS2 binds to and inhibits MDM2, which is an E3 ubiquitin that targets p53 for proteasomal degradation. Thus, activation of LATS2 also indirectly increases p53 levels. Centrosome amplification is known to promote LATS1/2 activation in an MST1/2-independent manner; however, the mechanisms leading to LATS1/2 activation in this context remain to be determined.
The upstream regulatory pathways that mediate LATS1/2 phosphorylation and activation are complex and not yet fully understood. In the classical signaling cascade (reviewed in (Pan, 2010, Yu and Guan, 2013, Zhao et al., 2010a, Zhao et al., 2010b, Zhao et al., 2011)), MST1 and MST2 kinases form heterodimers with the adaptor protein SAV1, which enhances MST1 and MST2 kinase activities. The MST/SAV1 complexes directly phosphorylate and partially activate LATS1/2 kinases. MST/SAV1 also phosphorylate MOB1, which enables it to bind to the autoinhibited regions of LATS1/2. MOB binding releases LATS1/2 of their inhibitory state and enables autophosphorylation within their activation loops. Together, the coordinated actions of MST1/2, SAV1, and MOB fully activate LATS1/2 (Figure 2). Loss of function of any of these components can inactivate the Hippo pathway, and all core members of this signaling pathway (MST1/2, LATS1/2, SAV1, and MOB) have tumor suppressive activities in mammals (Cai et al., 2010, Lee et al., 2010, McPherson et al., 2004, Nishio et al., 2012, Song et al., 2010, St John et al., 1999, Yabuta et al., 2007, Zhou et al., 2009b, Zhou et al., 2011).
In addition to the classical, linear MST1/2 signaling cascade, it is now recognized that regulation of YAP/TAZ can be achieved through MST1/2-independent processes in certain contexts. For example, disruption of the actin cytoskeleton and/or reduced RhoA activity, which occur upon cell detachment, serum starvation, tetraploidy, and contact inhibition, all activate LATS1/2 in an MST1/2-independent manner (Ganem et al., 2014, Mo et al., 2012, Wada et al., 2011, Yu et al., 2012, Zhao et al., 2012). This implies that additional regulatory mechanisms exist to activate LATS1/2 and inactivate YAP/TAZ, and new studies demonstrate that this regulation may involve other members of the Ste-20 family of kinases (Li et al., 2014, Zheng et al., 2015).
In addition to negatively regulating YAP/TAZ, activation of the Hippo pathway can also engage the p53 pathway. Phosphorylated, active LATS2 (but not LATS1) can bind and inhibit the E3 ubiquitin ligase mouse double minute 2 (MDM2), which normally targets p53 for destruction (Aylon et al., 2006). Consequently, LATS2 activation leads to p53 stabilization and the expression of downstream target genes that reinforce cell cycle arrest (e.g. cyclin dependent kinase inhibitor 1A (CDKN1A)). LATS2 itself is a p53 target gene, and its activation thus initiates a feedback loop to enforce Hippo pathway activity (Aylon et al., 2006). Activation of the Hippo pathway therefore limits cellular proliferation in at least two ways: by inactivating YAP/TAZ and by stabilizing p53 (Figure 2).
Activation of the Hippo Pathway Impairs the Proliferation of Cells with Extra Centrosomes
Extra centrosomes primarily arise from cytokinetic failures that give rise to tetraploid cells or from deregulation of mechanisms that control centriole overduplication (Figure 1). It has long been recognized that tetraploid cells activate the p53 pathway and fail to proliferate (Andreassen et al., 2001, Fujiwara et al., 2005, Ganem and Pellman, 2007). Interestingly, a recent study by Holland et al. demonstrates that centrosome amplification alone, independent of tetraploidy, similarly activates the p53 pathway and impairs long-term cell growth (Holland et al., 2012). This suggests that extra centrosomes, per se, can impair cell proliferation.
To induce extra centrosomes in non-transformed diploid cells, Holland et al. transiently overexpressed wild-type or degradation-resistant forms of the centriole duplication regulator Polo-like kinase 4 (PLK4) (Habedanck et al., 2005, Kleylein-Sohn et al., 2007). Clonogenic growth assays revealed that the resulting diploid cells, now containing numerous supernumerary centrosomes, exhibited severely impaired proliferation. Upon closer inspection, it was revealed that the cells had activated the p53 pathway. This stabilization of p53 was not simply an indirect effect of deregulated PLK4 kinase phosphorylating unidentified substrates, as expression of a non-degradable version of spindle assembly abnormal protein 6 homolog (SAS6) similarly led to centrosome amplification and reduced cell proliferation. Inactivation of the p53 pathway restored proliferation to the cells with extra centrosomes (Holland et al., 2012).
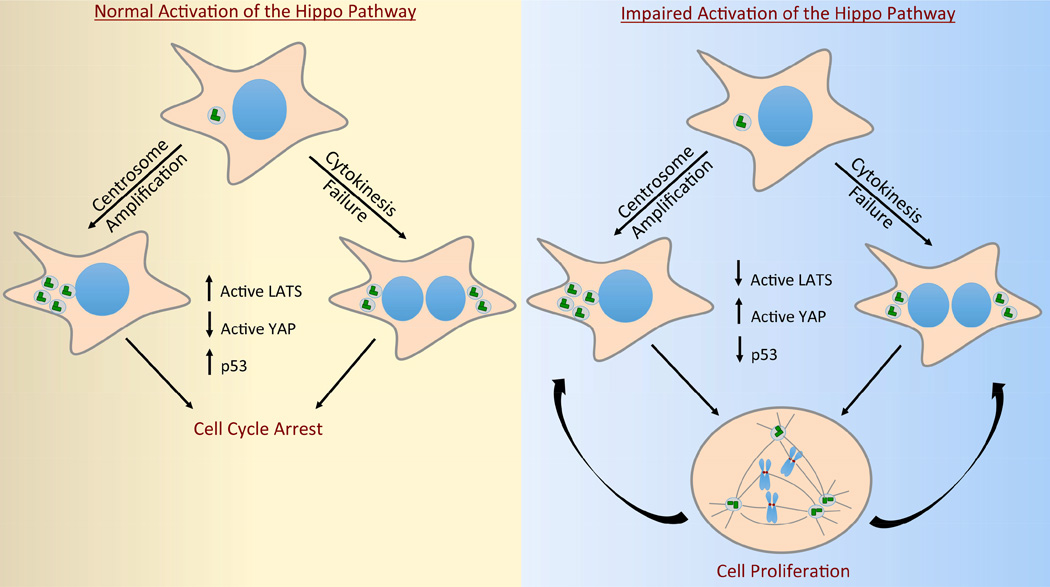
Recently, it has been shown that activation of the p53 pathway in both tetraploid cells and cells with centrosome amplification can be explained, at least in part, by activation of the Hippo pathway (Ganem et al., 2014). Extra-centrosomal cells display increased phosphorylation of LATS2 and subsequent inactivation of YAP (Figure 3). As described above, phosphorylated LATS2 binds to and inhibits MDM2, thus indirectly promoting the accumulation of p53 (Aylon et al., 2010). Importantly, depletion of LATS2 in cells with extra centrosomes mitigates p53 accumulation and activation of the p53 pathway (Ganem et al., 2014).
Figure 3. Inactivation of the Hippo pathway enables the proliferation of cells with extra centrosomes.
Centrosome amplification, either caused by centriole overduplication or cytokinesis failure, engages the Hippo pathway through increased activation of LATS1/2 kinases (left panel). Consequently, cells with extra centrosomes have decreased YAP activity and exhibit stabilization and accumulation of p53. These cells fail to proliferate (left panel). By contrast, cells that have functionally inactivated the Hippo pathway are capable of proliferation even if they possess supernumerary centrosomes (right panel). In addition to imparting cells with an increased tolerance for extra centrosomes, deregulation of many Hippo pathway core components can also promote further cytokinesis failure and centriole overduplication.
Defining the mechanism through which extra centrosomes activate the Hippo pathway remains an important area of investigation. Several recent studies have demonstrated that disruption of the actomyosin cytoskeleton and/or reduction in the activity of small G-protein RhoA have a major role in activating LATS in a MST-independent manner (Aragona et al., 2013, Dupont et al., 2011, Halder et al., 2012, Mo et al., 2012, Wada et al., 2011, Yu et al., 2012, Mana-Capelli et al., 2014). Indeed, cells with extra centrosomes exhibit a significant reduction in active RhoA (Ganem et al., 2014, Godinho et al., 2014). This reduction in RhoA is triggered, at least in part, by the indirect effects of increased microtubule nucleation from extra centrosomes. Dynamic microtubules are known to stimulate the activity of the small G-protein Rac1, and it has been reported that the increased microtubule nucleation from extra centrosomes hyperactivates Rac1 (Godinho et al., 2014). As active Rac1 antagonizes RhoA (Sander et al., 1999), increased Rac1 activity provides one molecular explanation for the observed loss of RhoA activity in cells with extra centrosomes.
An alternative possibility is that the centrosome may act as a scaffold that mediates LATS2 activation in the cell by physically localizing regulatory components of the Hippo pathway to a distinct subcellular space, and centrosome amplification would enhance this activation. Supporting this view, many core members of the Hippo pathway, including MST, MOB, SAV1 and LATS all localize to the centrosome throughout the cell cycle (Morisaki et al., 2002, McPherson et al., 2004, Nishiyama et al., 1999, Mardin et al., 2010, Guo et al., 2007, Wong et al., 2015, Hergovich et al., 2006, Hergovich et al., 2007, Hergovich et al., 2009). This localization is similar in principle to how LATS2 is activated at the plasma membrane, where it is recruited by neurofibromin 2 (NF2) so that it may more efficiently interact with MST/SAV complexes (Yin et al., 2013). The idea of the centrosome as a signal platform is not new; centrosomes are known to anchor hundreds of regulatory proteins that mediate activation of diverse cellular networks (Jackman et al., 2003, Sluder, 2005). It is interesting to speculate that the subcellular localization of LATS2 may dictate which regulatory proteins activate it. For example, while it is known that LATS2 is phosphorylated and activated by MST1/2 at the plasma membrane, centrosomal-localized LATS2 may be responsive to different regulatory proteins. To date, the kinases responsible for activating LATS in response to centrosomal defects remain unknown. It is possible that such kinases are specifically recruited or localized to centrosomes.
Regulation of Centrosome Number and Function by Hippo Pathway Components
In addition to acting as signaling conduits to regulate cell proliferation, many key members of the Hippo pathway moonlight as regulators of various aspects of centrosome biology. For example, MST1 and MOB1A/B play keys roles in centriole duplication (Hergovich et al., 2009). Depletion of either MST1 or MOB1A/B results in impaired centriole duplication, while overexpression of MOB1A/B promotes centriole amplification (Hergovich et al., 2009). Mechanistically, MOB1A/B bind to N-Myc downstream regulated 1 and 2 (NDR1/2) kinases, which facilitates their subsequent phosphorylation and activation by MST1 (Hergovich et al., 2009). Active NDR1/2 have been shown to be important for centriole duplication, though the mechanisms remain to be defined (Hergovich et al., 2009, Hergovich et al., 2007, Cook et al., 2014).
MST kinases, in complex with SAV1, also play an essential role in centrosome disjunction (Mardin et al., 2010). Replicated centrosomes are bound to one another during interphase by the linker proteins C-Nap1 and rootletin (Fry et al., 1998, Yang et al., 2006). Upon entry into mitosis, these linker proteins are phosphorylated by the kinase NIMA-related kinase (Nek2A), which leads to their disassembly (Bahe et al., 2005, Faragher and Fry, 2003). This process, termed centrosome disjunction, enables centrosome separation and bipolar mitotic spindle assembly. A complex of MST2/SAV1 mediates this process by directly phosphorylating and activating Nek2A (Mardin et al., 2010). Thus, loss-of-function of MST2 or SAV1 impairs normal centrosome disjunction and has the capacity to promote abnormal mitosis (Mardin et al., 2010).
LATS1 and LATS2 kinases also localize to centrosomes throughout the cell cycle, yet their function there remains poorly understood (Morisaki et al., 2002, McPherson et al., 2004, Nishiyama et al., 1999, Guo et al., 2007, Wong et al., 2015, Toji et al., 2004). Cells depleted of LATS1/2 show no defects in centriole duplication or centrosome disjunction. However, it has been reported that depletion of LATS2 impairs the recruitment of γ-tubulin to centrosomes, thus limiting microtubule nucleation during mitosis and hindering chromosome alignment (Abe et al., 2006).
Most strikingly, loss of Hippo pathway components indirectly deregulates centrosome numbers by disrupting normal cytokinesis. Knock-out studies in mice demonstrate that loss of Hippo pathway components LATS1, LATS2, MOB1A and MOB1B all promote cytokinesis failure and the production of tetraploid cells with extra centrosomes (McPherson et al., 2004, Nishio et al., 2012, St John et al., 1999, Yabuta et al., 2007). Tetraploid cells, by virtue of their extra centrosomes, are genetically unstable and are known to be significant contributors to tumorigenesis (Boveri, 1914, Davoli and de Lange, 2012, Fujiwara et al., 2005, Ganem et al., 2007, Zack et al., 2013). Combatting this potentially oncogenic effect of tetraploidy, it has been demonstrated that activation of the Hippo pathway in tetraploid cells limits their growth (Ganem et al., 2014). Thus, inactivation of Hippo pathway components not only promotes the generation of oncogenic tetraploid cells, but also imparts them with the ability to proliferate (Figure 3). Unsurprisingly, deletion of LATS1 and LATS2 is significantly more common in high-ploidy tumors than neardiploid tumors (Ganem et al., 2014).
Concluding Remarks
Both centrosome amplification and Hippo pathway inactivation are common in human tumors. New data suggest that these two phenotypes may be intertwined: Functional inactivation of the Hippo pathway may not only promote cytokinesis failure and/or centrosome overduplication, but also impart the resulting cells with an increased tolerance for the extra centrosomes. Understanding how cancer cells adapt to tolerate extra centrosomes has the potential to reveal new vulnerabilities that can be therapeutically exploited.
Acknowledgments
A.F.B is supported by a Biomolecular Pharmacology Training Grant from the NIH/NIGMS (5T32GM008541). N.J.G is the Aram V. Chobanian Assistant Professor of Medicine in the Shamim and Ashraf Dahod Breast Cancer Research Laboratories and is supported in part by grants from the Richard and Susan Smith Family Foundation, the Searle Scholars Program, the Karin Grunebaum Cancer Research Foundation, and the NIH/NCI (K99/R00 CA154531-01).
Abbreviations
- CIN
chromosome instability
- CDKN1A
cyclin dependent kinase inhibitor 1A
- LATS1, LATS2
Large tumor suppressor kinases 1 and 2
- Mats
Mps1 binder (MOB)
- Mps1
Monopolor spindle 1
- MST1, MST2
Mammalian sterile 20-like kinases 1 and 2
- NDR1/2
N-Myc downstream regulated 1 and 2 kinases
- Nek2A
NIMA-related kinase 2A
- NF2
neurofibromin 2
- SAS6
spindle assembly abnormal protein 6
- TAZ
transcriptional co-activator with PDZ-binding motif
- TEAD
transcriptional enhancer activation domain
- YAP
transcriptional co-activator yes-associated protein
References
- Abe Y, Ohsugi M, Haraguchi K, Fujimoto J, Yamamoto T. LATS2-Ajuba complex regulates gamma-tubulin recruitment to centrosomes and spindle organization during mitosis. FEBS Lett. 2006;580:782–788. doi: 10.1016/j.febslet.2005.12.096. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Avruch J, Zhou D, Fitamant J, Bardeesy N, Mou F, Barrufet LR. Protein kinases of the Hippo pathway: regulation and substrates. Semin Cell Dev Biol. 2012;23:770–784. doi: 10.1016/j.semcdb.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylon Y, Ofir-Rosenfeld Y, Yabuta N, Lapi E, Nojima H, Lu X, Oren M. The Lats2 tumor suppressor augments p53-mediated apoptosis by promoting the nuclear proapoptotic function of ASPP1. Genes Dev. 2010;24:2420–2429. doi: 10.1101/gad.1954410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R, Brunk K, Vinadogrova T, Peel N, Franz A, Khodjakov A, Raff JW. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- Boveri T. Zur Frage der Entstehung maligner Tumoren. Jena, Germany: Gustav Fisher Verlag; 1914. [Google Scholar]
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, Pan D. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Chavali PL, Putz M, Gergely F. Small organelle, big responsibility: the role of centrosomes in development and disease. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S, Okuda M, Mussman JG, Fukasawa K. Genomic convergence and suppression of centrosome hyperamplification in primary p53−/− cells in prolonged culture. Exp Cell Res. 2000;258:310–321. doi: 10.1006/excr.2000.4916. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–4225. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- Compton DA. Mechanisms of aneuploidy. Curr Opin Cell Biol. 2011;23:109–113. doi: 10.1016/j.ceb.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Hoa LY, Gomez V, Gomez M, Hergovich A. Constitutively active NDR1-PIF kinase functions independent of MST1 and hMOB1 signalling. Cell Signal. 2014;26:1657–1667. doi: 10.1016/j.cellsig.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155:948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S, Mccollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Stearns T. The centriole duplication cycle. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, Mayor T, Meraldi P, Stierhof YD, Tanaka K, Nigg EA. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Cornils H, Chiu SY, O'Rourke KP, Arnaud J, Yimlamai D, Thery M, Camargo FD, Pellman D. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131:437–440. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Pellman D. Causes and consequences of centrosome abnormalities in cancer. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Thery M, Pellman D. Oncogene-like induction of cellular invasion from centrosome amplification. Nature. 2014 doi: 10.1038/nature13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Tommasi S, Liu L, Yee JK, Dammann R, Pfeifer GP. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol. 2007;17:700–705. doi: 10.1016/j.cub.2007.02.055. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nature Reviews Molecular Cell Biology. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Kohler RS, Schmitz D, Vichalkovski A, Cornils H, Hemmings BA. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Lamla S, Nigg EA, Hemmings BA. Centrosome-associated NDR kinase regulates centrosome duplication. Mol Cell. 2007;25:625–634. doi: 10.1016/j.molcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Hergovich A, Stegert MR, Schmitz D, Hemmings BA. NDR kinases regulate essential cell processes from yeast to humans. Nat Rev Mol Cell Biol. 2006;7:253–264. doi: 10.1038/nrm1891. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH. Centrosomes and the art of mitotic spindle maintenance. Int Rev Cell Mol Biol. 2014;313:179–217. doi: 10.1016/B978-0-12-800177-6.00006-2. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Fachinetti D, Zhu Q, Bauer M, Verma IM, Nigg EA, Cleveland DW. The autoregulated instability of Polo-like kinase 4 limits centrosome duplication to once per cell cycle. Genes Dev. 2012;26:2684–2689. doi: 10.1101/gad.207027.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Guan K-L. The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012;23:785–793. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhang W, Wang B, Trinko R, Jiang J. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kango-Singh M, Nolo R, Tao C, Verstreken P, Hiesinger PR, Bellen HJ, Halder G. Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development. 2002;129:5719–5730. doi: 10.1242/dev.00168. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai ZC, Wei X, Shimizu T, Ramos E, Rohrbaugh M, Nikolaidis N, Ho LL, Li Y. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- Lambrus BG, Uetake Y, Clutario KM, Daggubati V, Snyder M, Sluder G, Holland AJ. p53 protects against genome instability following centriole duplication failure. J Cell Biol. 2015;210:63–77. doi: 10.1083/jcb.201502089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, Lim DS. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X, Cotton JL, Mao J, McCollum D, Jiang J, Czech MP, Xu L, Ip YT. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in Drosophila. Dev Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee J-S, Johnson RL. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub MR, Stearns T. Supernumerary centrosomes nucleate extra cilia and compromise primary cilium signaling. Curr Biol. 2012;22:1628–1634. doi: 10.1016/j.cub.2012.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-capelli S, Paramasivam M, Dutta S, McCollum D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell. 2014 doi: 10.1091/mbc.E13-11-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardin BR, Lange C, Baxter JE, Hardy T, Scholz SR, Fry AM, Schiebel E. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R. Centrosome amplification causes microcephaly. Nat Cell Biol. 2013;15:731–740. doi: 10.1038/ncb2746. [DOI] [PubMed] [Google Scholar]
- Mcpherson JP, Tamblyn L, Elia A, Migon E, Shehabeldin A, Matysiak-Zablocki E, Lemmers B, Salmena L, Hakem A, Fish J, Kassam F, Squire J, Bruneau BG, Hande MP, Hakem R. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J. 2002;21:483–492. doi: 10.1093/emboj/21.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J-S, Yu F-X, Gong R, Brown JH, Guan K-L. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisaki T, Hirota T, Iida S, Marumoto T, Hara T, Nishiyama Y, Kawasuzi M, Hiraoka T, Mimori T, Araki N, Izawa I, Inagaki M, Saya H. WARTS tumor suppressor is phosphorylated by Cdc2/cyclin B at spindle poles during mitosis. FEBS Lett. 2002;529:319–324. doi: 10.1016/s0014-5793(02)03360-4. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Stearns T. The centrosome cycle: Centriole biogenesis, duplication and inherent asymmetries. Nature Cell Biology. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio M, Hamada K, Kawahara K, Sasaki M, Noguchi F, Chiba S, Mizuno K, Suzuki SO, Dong Y, Tokuda M, Morikawa T, Hikasa H, Eggenschwiler J, Yabuta N, Nojima H, Nakagawa K, Hata Y, Nishina H, Mimori K, Mori M, Sasaki T, Mak TW, Nakano T, Itami S, Suzuki A. Cancer susceptibility and embryonic lethality in Mob1a/1b double-mutant mice. J Clin Invest. 2012;122:4505–4518. doi: 10.1172/JCI63735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama Y, Hirota T, Morisaki T, Hara T, Marumoto T, Iida S, Makino K, Yamamoto H, Hiraoka T, Kitamura N, Saya H. A human homolog of Drosophila warts tumor suppressor, h-warts, localized to mitotic apparatus and specifically phosphorylated during mitosis. FEBS Lett. 1999;459:159–165. doi: 10.1016/s0014-5793(99)01224-7. [DOI] [PubMed] [Google Scholar]
- Oka T, Mazack V, Sudol M. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantalacci S, Tapon N, Leopold P. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- Paramasivam M, Sarkeshik A, Yates JR, 3rd, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468:321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancati G, Pavelka N, Fleharty B, Noll A, Trimble R, Walton K, Perera A, Staehling-Hampton K, Seidel CW, Li R. Aneuploidy underlies rapid adaptive evolution of yeast cells deprived of a conserved cytokinesis motor. Cell. 2008;135:879–893. doi: 10.1016/j.cell.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Cimini D, Cameron LA, Deluca JG. Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci. 2005;360:553–568. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander EE, Ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J Cell Biol. 1999;147:1009–1022. doi: 10.1083/jcb.147.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science. 2011;333:1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G. Two-way traffic: centrosomes and the cell cycle. Nat Rev Mol Cell Biol. 2005;6:743–748. doi: 10.1038/nrm1712. [DOI] [PubMed] [Google Scholar]
- Sluder G. One to only two: a short history of the centrosome and its duplication. Philos Trans R Soc Lond B Biol Sci. 2014;369 doi: 10.1098/rstb.2013.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, Wang C-Y, Gao B, Jiang J, Yang Y. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature. 2010;464:436–440. doi: 10.1038/nature08803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St John MA, Tao W, Fei X, Fukumoto R, Carcangiu ML, Brownstein DG, Parlow AF, Mcgrath J, Xu T. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber D, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–672. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188:369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci U S A. 2011;108:17974–17978. doi: 10.1073/pnas.1109720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toji S, Yabuta N, Hosomi T, Nishihara S, Kobayashi T, Suzuki S, Tamai K, Nojima H. The centrosomal protein Lats2 is a phosphorylation target of Aurora-A kinase. Genes Cells. 2004;9:383–397. doi: 10.1111/j.1356-9597.2004.00732.x. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006a;18:74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Tsou MFB, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006b;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Wada K-I, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, Mitchell BJ, Desai A, Gahman TC, Shiau AK, Oegema K. Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science. 2015;348:1155–1160. doi: 10.1126/science.aaa5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- Yabuta N, Okada N, Ito A, Hosomi T, Nishihara S, Sasayama Y, Fujimori A, Okuzaki D, Zhao H, Ikawa M, Okabe M, Nojima H. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol Biol Cell. 2006;17:1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F-X, Guan K-L. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F-X, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu X-D, Mills GB, Guan K-L. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, Sougnez C, Gabriel SB, Hernandez B, Shen H, Laird PW, Getz G, Meyerson M, Beroukhim R. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Guan KL. Hippo signaling at a glance. J Cell Sci. 2010a;123:4001–4006. doi: 10.1242/jcs.069070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan K-L. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010b;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang C-Y, Yu J, Guan K-L. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Tumaneng K, Guan K-L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai Z-C, Guan K-L. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang W, Liu B, Deng H, Uster E, Pan D. Identification of Happyhour/MAP4K as Alternative Hpo/Mst-like Kinases in the Hippo Kinase Cascade. Dev Cell. 2015 doi: 10.1016/j.devcel.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park J-S, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009a;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, Bardeesy N. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009b;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, Avruch J. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312–E1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]