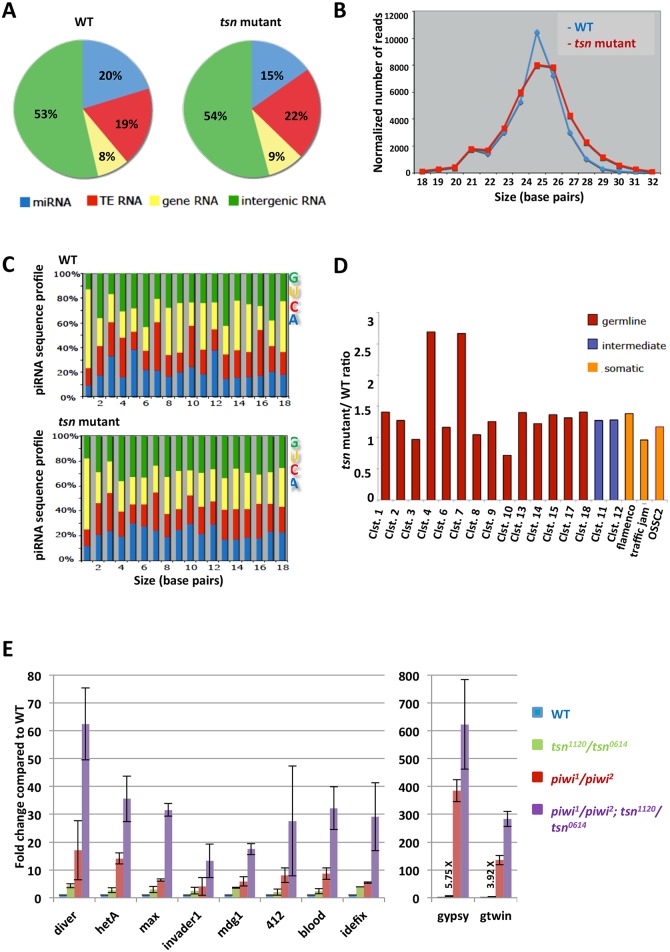
Fig 7. TSN shows only subtle effects on piRNAs and transposon silencing.
(A) Small RNA composition in the WT (left) and tsn1120/tsn0614 mutant (right) testes. TE RNA, small RNAs derived from transposon regions; gene RNA, small RNAs derived from gene-coding regions; intergenic RNA, small RNAs derived from intergenic regions. (B) Size distribution of small RNAs (excluding miRNAs and fragments of long cellular RNAs such as tRNAs and rRNAs) in the WT (blue) and tsn1120/tsn0614 mutant (red) testes. The level of 25-nt small RNAs was slightly decreased in the tsn mutants. (C) Nucleotide compositions of small RNAs (excluding miRNAs and fragments of long cellular RNAs such as tRNAs and rRNAs) in the WT (top) and tsn1120/tsn0614mutant (bottom) testes. The 1U-bias piRNA signature and nucleotide composition were not affected in tsn mutants. (D) Relative abundance of small RNAs derived from 19 previously reported piRNA clusters in tsn mutant testes compared to the WT testes are shown. Small RNAs uniquely mapped to these clusters were used for this analysis. These clusters are categorized into three groups according to the expression preference of their piRNAs [39–41], with the germline-enriched class colored in red, the intermediate class colored in blue, and the soma-enriched class colored in orange. No strong impact of tsn mutations on these piRNA clusters was observed (see text). (E) qRT-PCR was performed to determine the expression of multiple transposons, relative to rp49, in tsn1120/tsn0614 mutant testes. Transcript levels from the WT testes were set as 1, and fold-changes are indicated. Error bars represent mean ± standard error of the mean (N = 3). tsn mutants showed very moderate defects in transposon silencing (see text).