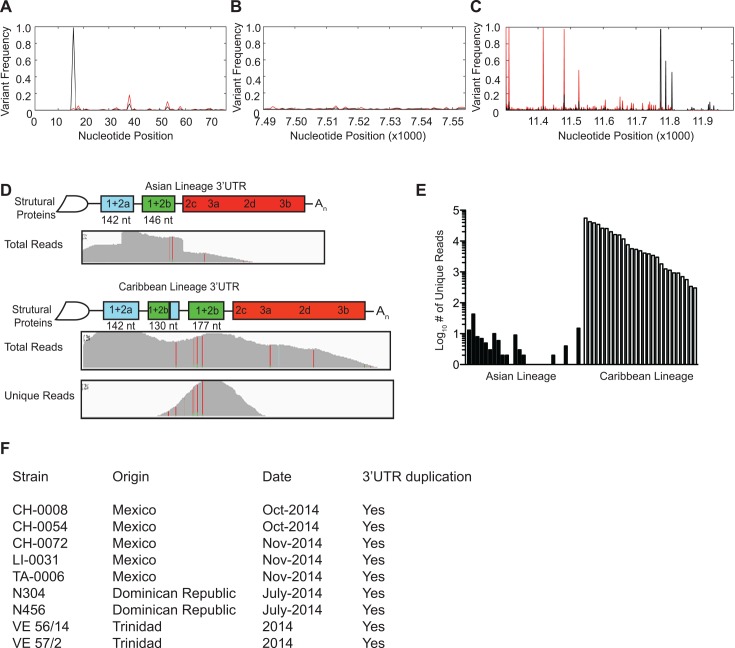
Fig 5. Analysis of minority variants in noncoding genome regions and identification of a novel duplication in the 3’UTR.
A-C. Minority variants present in the 5’UTR, genome nucleotides 1–76 (A), subgenomic promoter, genome nucleotides 7490–7554 (B), and 3’UTR, genome sequence 11302–11944 (C). Graphs represent the mean variant frequency (black) and variant range (red) at each nucleotide. D. Schematic and alignment of the published Asian strain and novel Caribbean strain containing a 177 nucleotide duplication of the 3’ end of 1+2a and complete 1+2b in the 3’UTR. “Total reads” show the read coverage of all reads aligning to the two lineage references. E. Quantification of the total “unique reads”. Unique reads show the read coverage of all reads where the alignment to the Caribbean reference is superior to the alignment of the Asian Lineage. F. The presence of the 3'UTR duplication was confirmed in clinical samples obtained from Mexico [18], Dominican Republic and Trinidad.