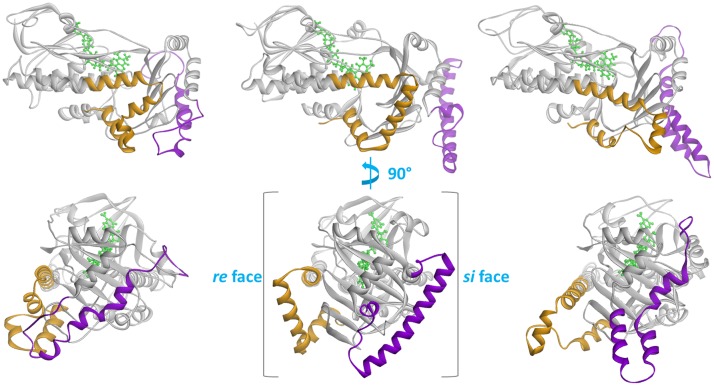
Fig 3. Comparison of the three Coq6p homology models.
The Coq6p enzyme secondary structure is shown in cartoon, while the FAD cofactor is shown in green stick. Structurally divergent Coq6p secondary structure elements are highlighted in orange (the C-terminus) and purple (the Coq6-family insert). From left to right: Coq6p_I-TASSER model, Coq6p_ROBETTA model, and the Coq6p_MODELLER model. The isoalloxazine ring plane of the FAD co-factor has two faces, named si and re, with the relevant stereocenter for naming the faces being the C2’ carbon of the ribityl chain. Each face of the FAD’s ring is used here to designate the half of the Coq6p enzyme extending from the si and re faces of the FAD binding pocket to the edge of the protein. The top row shows the models from the re face, while the bottom row is rotated 90 degrees about the vertical to show the exterior of the beta sheet domain, which bears the insert.