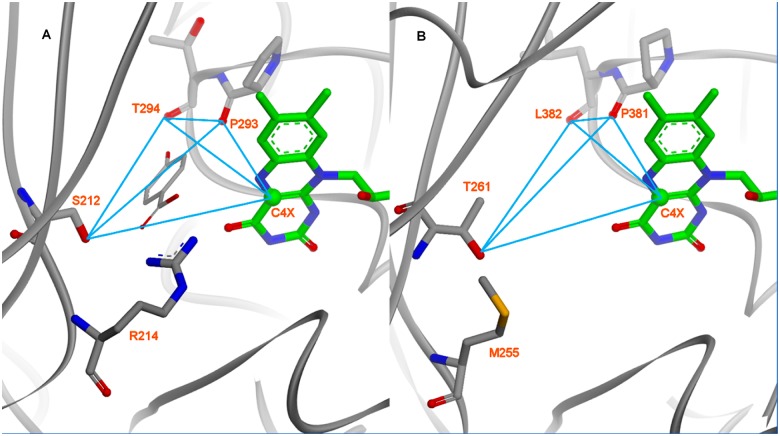
Fig 5. Comparison of pHBH’s active site crystal structure with the same region in the Coq6p model (here, the Coq6p_MODELLER model).
The active site interatomic distances used for the scoring function-based substrate docking are shown as blue lines (see Methods). A): The co-crystal structure of para-hydroxybenzoate in pHBH (1PBE) provides coordinates of a structurally and functionally homologous enzyme-substrate-co-factor complex. In order to capture the active-site geometry compatible with substrate binding, a receptor-based geometric descriptor is derived from the enzyme’s hydrogen bonding partners with the substrate. B): Residues and atoms used in the geometric descriptor derived for pHBH are mapped onto homologous residues and atoms in the Coq6p catalytic site.