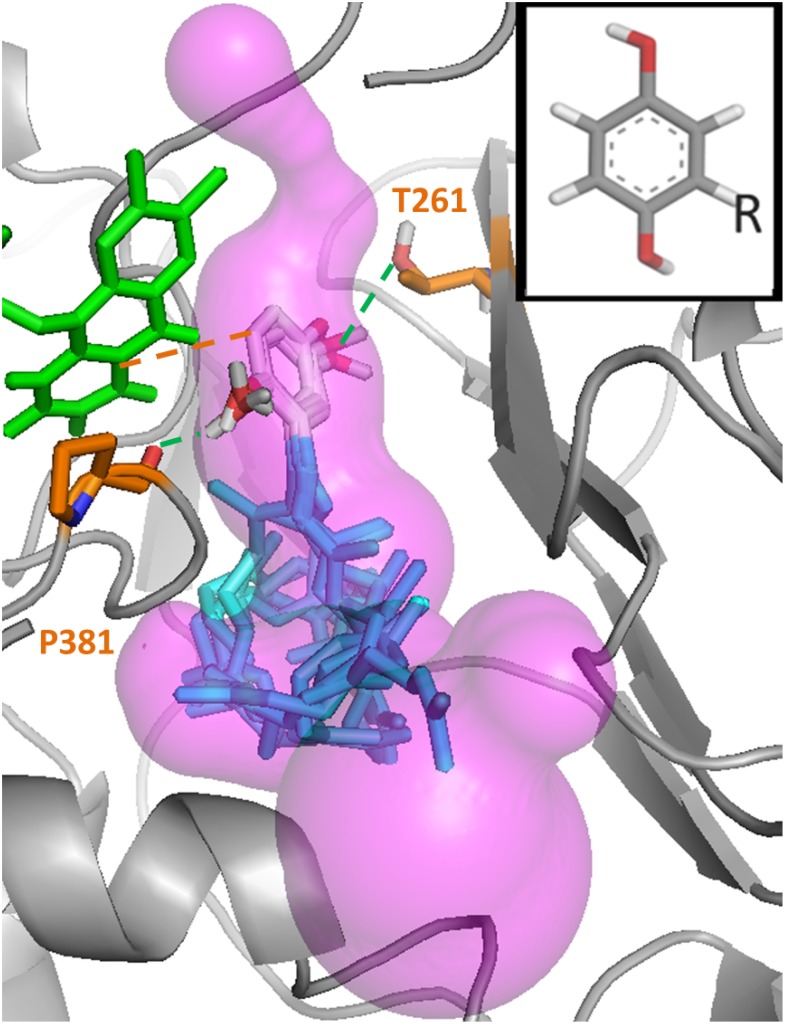
Fig 7. Recurrent catalytically plausible poses of the 3-hexaprenyl-4-hydroxyphenol model substrate in the re face tunnel 1 of the Coq6p_MODELLER model.
The enzyme conformation for the substrate docking calculations was selected with the active site geometric descriptor scoring-function derived from 1PBE[40] (see Methods). The FAD co-factor is represented in green stick and the substrate in grey (aromatic head) and blue (isoprenyl chain). Several substrate poses are superimposed within the accessible volume (transparent pink) delimited by the re face substrate access tunnel 1. The substrate’s C1 and C6 hydroxyl groups can form hydrogen bonds (green dashed lines) with the backbone oxygen of P381 and the side chain oxygen of T261. One pose is also illustrated that shows a distance between the substrate’s C5 carbon (the hydroxylation target) and the FAD’s C4X (which bears the reactive peroxo group) of 4.7Å (dashed orange line), similar to the homologous distance observed in the 1PBE structure of 4.3Å.